All talks will presented live regardless if you are participating in-person or virtually. While most presenters will be in-person, those unable to travel will be presented live through the conference platform and streamed into the venue presentation room.
As you plan your participation as a presenter please find below details to assist you with your specific presentation type at the conference:
- How to Join the Session as a Virtual Speaker
- In-person Talk Computer Details
- Live Talks and Pre-recorded Videos for the Content Library
- Poster Presentation
- Recorded Presentation Guidelines
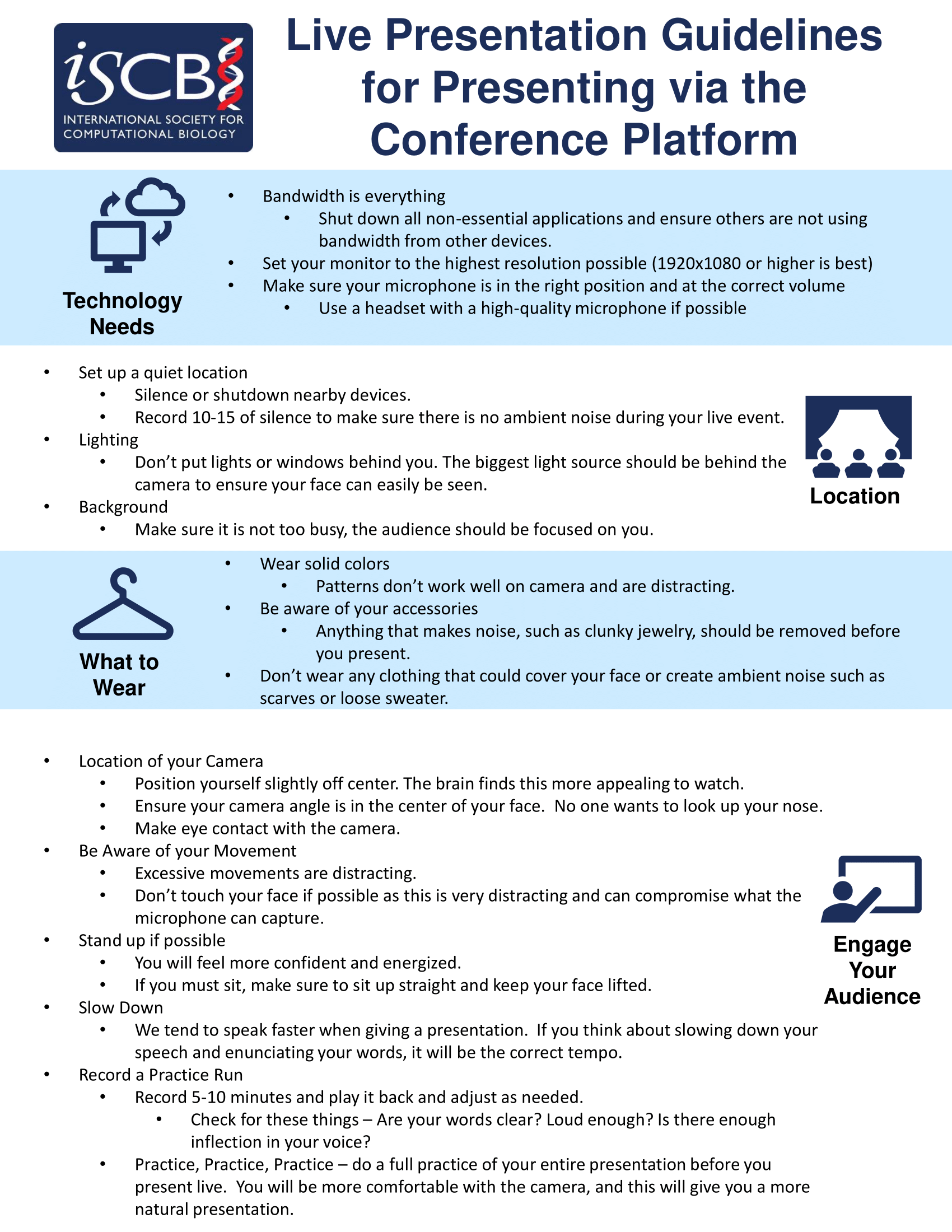
- Live Presentation Guidelines
- How to Record a PowerPoint Presentation
- Hybrid Conference Optimal Conditions
- Uploading your Talk
- Uploading your Poster
You can also find additional training videos here: https://help.junolive.com/exhibitors-speakers/training-videos-for-exhibitors-speakers-moderators
To join the session:
- log in to the virtual platform (https://iscb.junolive.co/) using the email you registered for the conference with.
- It is important that you use the same email as permissions on the virtual site are tied to your email
- Once logged in navigate to your talk
- There are multiple paths to your talk, for example searching through the Scientific Programme or via your profile page
- Normally clicking on your talk will bring up the details about your talk. 30 minutes before the session where your talk is scheduled, that link will instead take you to the video conferencing system. This is why the email permissions are important.
We ask that you join as early as you are able within the 30 minute window to provide ample time to troubleshoot any issues that may arise. Please note, this is 30 minutes before the session containing your talk, not 30 minutes before your talk
NOTE: sharing the screen with the virtual audience means presenter mode cannot be used unless you have a second monitor and configure it beforehand. For more information about screen sharing with a single monitor visit this page: https://help.junolive.com/exhibitors-speakers/screen-sharing-with-a-single-monitor
If you need assistance joining the room there is a help chat available that will attempt to troubleshoot and connect you to live assistance. You can also contact This email address is being protected from spambots. You need JavaScript enabled to view it.
For those presenting in-person please note you must use the supplied computer due to the connection process with the JUNO virtual platform. You should pre-load your presentation on the in-room computer prior to your event.
Computer Specifications:
- All laptops will run Microsoft Windows 10 and include PowerPoint, Adobe PDF, and Microsoft Office. PDFs will not be able to have the fullscreen shown on the podium computers. As such, powerpoint is highly recommended
- All laptops will have Microsoft Office 365, compatible with the old version from 2016. There should be no compatibility issues for presentations created with PowerPoint 2019 or 2021.
- If video will be a part of your presentation, it is highly recommended for it to be inserted using the “Video on my PC” option within PowerPoint. Save/include the video file included in the same folder as your presentation. It is not recommended to use the “Online Video” option. Presentation times will not be extended for troubleshooting technical difficulties for videos that do not load.
- Custom/non-standard Windows fonts are not recommended or supported. If utilized, ensure that the font(s) are fully embedded within your presentation file.
- Apple Keynote and Google Slides are not supported.
- Slide size should be set to “Widescreen (16:9)” within PowerPoint (this is the default).
- Projector Information: 16:9 Aspect Ratio / 1920x1080 resolution. Screen size will vary depending upon the room.
All abstract presenters, invited speakers, poster presenters, and proceedings presenters are required to provide a pre-recorded talk for the virtual platform library in advance of the conference. The pre-recorded talk will be available shortly after the live presentation to provide faster and more seamless access to the conference content for virtual participants.
All poster presenters, including those presenting in-person, will upload a short 5-7 minute video to the virtual conference platform site along with a PDF of their poster. Those presenting a poster and a talk on the same topic must provide two sets of files, one for the talk one for the poster. A talk video cannot be used for a poster presentation as the allowed time is shorter.
Pre-recorded talks for the platform library should be prepared ensuring you upload a .mp4 file. Videos should be recorded in 720p - 1080p and there is no limit on video file size. Poster PDFs must not exceed 10Mb. Presenters will upload their recordings directly through the conference platform site after logging in as a registrant.
PLEASE note once submitted your video cannot be updated, though it can be removed and replaced.
- Save your presentation as .mp4 file
- Ensure you use the filename(s) provided to you in the email sent upon submission of the Confirmation of Participation. This email will be sent from This email address is being protected from spambots. You need JavaScript enabled to view it., be sure it is whitelisted so as not to miss any communications from ISCB
- If you are presenting a poster in addition to a talk you must prepare a second video for the poster as the allowed time is shorter (5-7 minutes). The video for your talk cannot be used for your poster presentation. See the poster instructions below for more details
- Recording may or may not include the presenter in the recording, this is at the presenter's discretion. Should the presenter elect to appear on camera in your recording, we suggest you wear the same outfit the day of your presentation.
- Ensure you prepare your talk based on the length of time specified within the acceptance notification :
The following links can be of assistance for planning your presentation(s):
Some additional helpful tips on planning your recorded talk are available below. We suggest that if you are including a visual image of yourself in the recording you consider wearing the same outfit for the live Q&A.
Turn on live captions using the Google Chrome browser. Download Chrome here if you haven’t already. Remember, you’ll always need to access the event website in Chrome for these captions to work. More details can be found at https://help.junolive.com/attendees/turn-on-live-captions
Presenters of a poster will upload your poster and video via the conference platform with the following:
- Presentation abstract
- PDF of your poster, no larger than 10Mb
- 5 to 7 minute poster presentation talk as .mp4 file (Maximum 7 minutes)
- Videos should be recorded in 720p - 1080p and there is no limit on video file size
Poster presenters will be able to:
- direct message with attendees
- create ad hoc "video demo rooms"
- Q&A with the poster presenter via public comments
Some helpful tips on planning your recorded talk are available below.
Presenting your poster in a lighting style format using the PechaKucha or Ignite talks presentation style or a single slide or PDF is an option for presenters.
Here is an example of a presentation: https://youtu.be/rbLbb7eOao8
Posters can be no larger than 46"x46". Only in-person presenters need to print a poster.
![]()
How to record a power point presentation
For the best experience please consider the following in preparation of the conference:
- Preferred browser is Chrome
- Hardwire your computer vs wifi
- Turnoff other browsers, background programs and eliminate other internet devices being used if possible
A pre-recording version of your talk is required as a back-up precaution, should anything prevent you from being able to present in-person. We would rather play your recorded talk then have 'dead time' within the programme. Additionally, the pre-recorded talk will be available shortly after the live presentation to provide faster and more seamless access to the conference content for virtual participants. To ensure all required presentation materials are available be sure to follow these steps to upload your files
- Once the window for you to upload your poster opens you will be sent a "You have been added as an Admin" email from ISCB
- Log in to the conference virtual platform using the email you registered with
- Navigate to your presentation. You can do so with the link in the "You have been added as an Admin" email, or you can navigate to your talk using the "Scientific Programme" on the virtual platform
- Once in the page for your talk you will need to scroll down to click on your name, then visit page to go to your speaker profile
- Click the edit button on your speaker profile
- Use this page to add a photo and speaker bio (OPTIONAL) and be sure to click the "Save Changes" button after any updates.
- To upload your mp4 navigate to the "Meta Data" tab and click the "Add Meta Data" button
- Select "Upload MP4" from the type menu
- Click Choose a file then navigate to the video on your computer. Be sure to use the filename provided upon submission of the Confirmation of Participation. Using the incorrect filename may result in the file not being properly displayed.
- Confirm you have the right video and a pop-up will appear showing you progress of the upload. Once completed the "Cancel" button will change to "OK"
- Click "Save Changes"
The following video tutorial from ISMB 2022 will walk you through the same steps
A digital version of your poster as well as an accompanying flash talk ensures all posters are viewable to both in-person and virtual attendees thus providing more exposure of your work. Failure to upload a digital version of your poster will revoke your in-person poster acceptance. To ensure all required presentation materials are available be sure to follow these steps to upload your files
- Once the window for you to upload your poster opens you will be sent a 'You have been added as an Admin' email from ISCB with a link that will take you directly to the front end of your poster booth
- Additionally, you'll receive a notification on in your profile on the conference site that will direct you to the same location as the email
- Information tab = OPTIONAL Intro, Description, and Image update
- Rotators - this is where you'll upload the mp4 of your presentation. The steps to do so are:
- Click "Add rotator"
- Give the rotator a title
- Type - use the drop down to find "Video Upload mp4"
- Upload file - this will open to search your computer for the file. Be sure to use the filename provided upon submission of the Confirmation of Participation. Using the incorrect filename may result in the file not being properly displayed
- Once uploading, don't navigate to another page - you may use a separate tab while the video is uploading, though
- Once completed, the "CANCEL" button will update to "OK". Click "OK" and then save changes
- Resources - this is where you'll upload your pdf. This process is the same as the mp4
- Add a title (required)
- Description is optional and not shown on the site
- Upload your file using 'Upload File'. Again, this will allow you to search your local computer for the file you'd like to upload
- Be sure to use the filename provided upon submission of the Confirmation of Participation. Using the incorrect filename may result in the file not being properly displayed
- File must be less than 10 MB - this limit only applies to the pdf
- The same downloading window will appear, and you'll click Complete when the file has been uploaded.
- Meta Data - this is where you may add in any personal links/ways to contact you (Optional)
- Greeting - This will populate next to the rotator on the page (Optional)
The following video tutorial from ISMB/ECCB 2023 will walk you through the same steps
Tuesday - Day 1 – November 28, 2023 (RSG)
*Schedule subject to change without notice
| Jump to: Wednesday | Thursday | ||
|---|---|---|
| Start Time | End Time | Session |
| 8:30 AM | 5:30 PM | Registration |
| 8:30 AM | 9:00 AM | Badge pick up, upload talks and Coffee |
| 9:00 AM | 10:00 AM | RSG Welcome and KEYNOTE 1 - David Kelley |
| 10:00 AM | 10:30 AM | Session 1 |
| 10:30 AM | 10:45 AM | Bio Break |
| 10:45 AM | 11:30 AM | Session 2 |
| 11:30 AM | 1:30 PM | Poster Session with lunch |
| 1:30 PM | 2:15 PM | KEYNOTE 2 - Jingyi Jessica Li |
| 2:15 PM | 3:30 PM | Session 3 |
| 3:30 PM | 4:00 PM | Coffee break with snacks |
| 4:00 PM | 4:30 PM | Session 4 |
| 4:30 PM | 5:15 PM | KEYNOTE 3 - David Van Valen |
Wednesday - Day 2 – November 29, 2023 (RSG)
*Schedule subject to change without notice
| Jump to: Tuesday | Thursday | ||
|---|---|---|
| Start Time | End Time | Session |
| 8:30 AM | 5:30 PM | Registration |
| 8:30 AM | 9:00 AM | Badge pick up, upload talks and Coffee |
| 9:00 AM | 10:00 AM | RSG Welcome and KEYNOTE 4 - Jean Fan |
| 10:00 AM | 10:30 AM | Session 5 |
| 10:30 AM | 10:45 AM | Bio Break |
| 10:45 AM | 11:30 AM | Session 6 |
| 11:30 AM | 1:30 PM | Poster Session with lunch |
| 1:30 PM | 2:15 PM | KEYNOTE 2 - Remo Rohs |
| 2:15 PM | 3:15 PM | Session 7 |
| 3:15 PM | 3:45 PM | Coffee break with snacks |
| 3:45 PM | 5:00 PM | Session 8 |
Thursday - Day 3 – November 30, 2023 (DREAM)
| Jump to: Tuesday | Wednesday | ||
|---|---|---|
| Start Time | End Time | Session |
| 8:30 AM | 5:30 PM | Registration |
| 9:00 AM | 9:30 AM | Welcome to DREAM Day |
| 9:30 AM | 10:00 AM | Coda TB Challenge |
| 10:00 AM | 10:10 AM | Short Break |
| 10:10 AM | 10:40 AM | scRNA-seq and scATAC-seq Data Analysis DREAM Challenge |
| 10:40 AM | 11:10 AM | Overview of openchallenges.io challenge registry |
| 11:10 AM | 11:40 AM | Upcoming DREAM challenges |
| 11:40 AM | 1:10 PM | Lunch and Posters |
| 1:10 PM | 2:10 PM | Keynote - Paul Spellman |
| 2:10 PM | 3:00 PM | FINRISK Heart Microbiome DREAM Challenge |
| 3:00 PM | 3:30 PM | Coffee Break with snacks |
| 3:30 PM | 5:00 PM | Xcelerate RARE: A Rare Disease Open Science Data DREAM Challenge |
 Ferhat Ay, PhD
Ferhat Ay, PhD
La Jolla Institute for Immunology
Ferhat Ay is the Institute Leadership Associate Professor of Computational Biology at the La Jolla Institute for Immunology (LJI) with adjunct appointments at the UC San Diego School of Medicine, Moores Cancer Center, Institute for Genomic Medicine and Bioinformatics and Systems Biology PhD program. Prior to LJI, he was a Research Assistant Professor at Northwestern University (2015) and a CRA Computing Innovation Fellow in William Noble's lab in the Department of Genome Sciences at the University of Washington (2011-2014). Ferhat completed his Ph.D. in Computer Science at the University of Florida (2007-2011) and his B.S. degrees in Computer Engineering and Mathematics both from Middle East Technical University (METU), Turkey, in 2007. His primary research areas are bioinformatics, computational biology, epigenomics, and regulatory genomics. Ferhat and his lab developed computational methods for studying chromatin conformation capture data (Hi-C, HiChIP), including highly utilized tools such as FitHiC, FitHiChIP, Mustache, HiCNV/HiCTrans and dcHiC. He serves as an Associate Editor for PLoS Computational Biology, Journal of Computational Biology, and as an Editorial Board Member for Genome Biology, and in several different organizational roles for ISCB conferences, including ISMB, RECOMB and RSGDREAM.
 Elise Blaese, MS, MBA
Elise Blaese, MS, MBA
IBM
Elise Blaese is a Senior Project Manager for IBM Research. She supports the work of the IBM Research Healthcare and Life sciences team. She is also a DREAM Director, having worked on outreach, marketing, logistics and management of DREAM Challenges and the Annual DREAM conference since 2011. Elise joined IBM Research in 2008 where she works to find research opportunities for IBM researchers and our partner universities with the NIH. Before joining Research Elise work in IBM’s internal healthcare startups in solutions development and marketing for five years. Prior to IBM, Elise worked for NASA for 10 years in the Space Life Sciences research program where she was a project manager for the Life Sciences Data Archive (LSDA). Elise also worked in payload Mission Control for four Spacelab missions in the early 90’s.
 Jason Ernst, PhD
Jason Ernst, PhD
University of California Los Angeles
Jason Ernst is a Professor of Biological Chemistry, Computer Science, and Computational Medicine at UCLA. Prior to joining the UCLA faculty, he was a postdoctoral fellow with Manolis Kellis in the MIT Computer Science and Artificial Intelligence Laboratory and affiliated with the Broad Institute. In 2008, Jason completed a PhD advised by Ziv Bar-Joseph in the Machine Learning Department and School of Computer Science at Carnegie Mellon University. Jason also earned BS degrees in Computer Science and Mathematics from the University of Maryland College Park. He has developed a number of widely used bioinformatics methods and software include ChromHMM for chromatin state modeling and genome annotation and STEM and DREM for the analysis of time series gene expression data. He is a member of the editorial board at Genome Research and has been a program co-chair for the Regulatory Genomics Special Interest Group meeting at ISMB. He is a recipient of a NIH-Avenir Award, NSF CAREER Award, Sloan Fellowship, and a John H. Walsh Young Investigator Research Prize.
 Pablo Meyer, PhD
Pablo Meyer, PhD
IBM
Pablo Meyer is Manager of the Biomedical Analytics and Modeling group at IBM research, Director of DREAM challenges and as such has been working on developing models and applying AI to biological/biomedical problems, Systems Biology, Olfaction and Disease.
 Sushmita Roy, PhD
Sushmita Roy, PhD
University of Wisconsin-Madison
Sushmita Roy is a Professor at the Biostatistics and Medical Informatics Department and a faculty at the Wisconsin Institute for Discovery, University of Wisconsin, Madison. Her research lies at the intersection of machine learning and regulatory genomics. Her group develops and applies computational methods for identifying regulatory networks that exist in living cells and examines their dynamics across different biological contexts. She works closely with experimentalists who study a variety of biological processes ranging from infectious disease, cell fate specification, host microbe interactions, evolution of tissue-specific gene expression that all have a shared goal to understand the underlying regulatory network. Dr. Roy is a recipient of an Alfred P. Sloan Foundation Fellowship, an NSF CAREER award and a James McDonnell foundation scholar award.
Venue
 Conference will take place in the Palisades Room of Carnesale Commons at UCLA
Conference will take place in the Palisades Room of Carnesale Commons at UCLA
The address is: 251 Charles E Young Drive West, Los Angeles, CA 90095
Accomodations
UCLA Guest House
 When you stay at the UCLA Guest House, the goal of our friendly, knowledgeable staff is to help you feel right at home. We've taken care of all the important details that make your stay a pleasure — luxurious beds and linens, attentive housekeeping, upscale bath amenities, complimentary wireless Internet throughout the hotel, and convenient on-site shuttle services.
When you stay at the UCLA Guest House, the goal of our friendly, knowledgeable staff is to help you feel right at home. We've taken care of all the important details that make your stay a pleasure — luxurious beds and linens, attentive housekeeping, upscale bath amenities, complimentary wireless Internet throughout the hotel, and convenient on-site shuttle services.
Guests can enjoy complimentary continental breakfast each morning, self-serve coffee and tea 24 hours a day, and a selection of newspapers provided daily in the lobby. Hotel patrons will enjoy the convenience of being close to campus landmarks and attractions, including beautiful gardens and museums.
- Room Rate: $229.00-$239.00 per night based on room type with no taxes or additional fees
- Reservations can now be made directly with the Guest House, while rooms last by using this link:
UCLA Luskin Conference Center Hotel
 Guest rooms at the UCLA Meyer and Renee Conference Center’s hotel were designed with every comfort and convenience in mind. Each room is imbued with a warm ambiance that encourages relaxation, so guests can power down from a long day and wake rejuvenated to embrace an exciting tomorrow. Peace of mind comes easy the moment one enters, as the room’s motion sensor thermostat creates the perfect climate, all while preserving precious energy resources.
Guest rooms at the UCLA Meyer and Renee Conference Center’s hotel were designed with every comfort and convenience in mind. Each room is imbued with a warm ambiance that encourages relaxation, so guests can power down from a long day and wake rejuvenated to embrace an exciting tomorrow. Peace of mind comes easy the moment one enters, as the room’s motion sensor thermostat creates the perfect climate, all while preserving precious energy resources.
The Luskin Conference Center is an enticing place to stay near UCLA, and one of the finest hotels in Westwood. Located near the UCLA Medical Center, its 254 spacious hotel rooms are appointed with one king bed or two queens, all with deluxe Pima cotton linens and super soft mattress toppers and pillows for a superb night’s rest. All rooms feature earth-friendly, premium toiletries and plush bathrobes so guests can await inspiration while wrapped in comfort. Through our partnership with Clean the World, we ensure that no resource goes to waste, as we donate and recycle all leftover toiletries; that unused soap may very well reach a person in need on the other side of the globe.
- Room Rate offered is 15% off the best available rate with our special link:
 Jean Fan
Jean Fan
John Hopkins University
USA
Jean Fan is a member of the faculty of Biomedical Engineering in the Center for Computational Biology at Johns Hopkins University. Her research team, the JEFworks lab, is interested in understanding the molecular and spatial-contextual factors shaping cellular identity and heterogeneity. She develops new open-source computational software for analyzing spatially-resolved omics data that can be tailored and applied to diverse tissue types and biological systems. Dr. Fan is also the founder, director, and lead software developer for the non-profit organization CuSTEMized, which provides personalized STEM picture storybooks to encourage young girls to see themselves as scientists. Dr. Fan was previously an NCI F99/K00 post-doctoral fellow in the lab of Dr. Xiaowei Zhuang at Harvard University. She received her PhD in Bioinformatics and Integrative Genomics at Harvard under the mentorship of Dr. Peter Kharchenko at the Department of Biomedical Informatics and in close collaboration with Dr. Catherine Wu at the Dana-Farber Cancer Institute. The impact of Dr. Fan's work has been recognized by several awards and honors, including the Forbes 30 Under 30, the Nature Research Award for Inspiring Science, and the NSF CAREER Award.
 David Kelley
David Kelley
Calico Life Sciences
USA
David Kelley is a Principal Investigator at Calico Life Sciences, where he studies gene regulation in aging. He completed his PhD in computer science at the U. of Maryland with Steven Salzberg, working on algorithms for genome assembly and gene prediction. Subsequently, he worked as a postdoc with John Rinn at Harvard on computational analyses of long noncoding RNA regulation and function. David developed the deep convolutional neural network toolkits Basset, Basenji, and Enformer to predict tissue-specific regulatory activity of DNA sequences. His group is interested in fully elucidating the mechanisms by which a regulatory environment acts on whole chromosomes to determine the expression of every gene. At Calico, he’s applying such models to study how disturbed regulatory connections influence the increased rate of disease onset over human lifespans.
 Jingyi Jessica Li
Jingyi Jessica Li
UCLA
USA
Jingyi Jessica Li is a Professor in the Department of Statistics at the University of California, Los Angeles, where she holds secondary appointments in the Departments of Biostatistics, Computational Medicine, and Human Genetics. She leads a research group called the Junction of Statistics and Biology, which focuses on developing interpretable statistical methods for biomedical data analysis.
Jessica received her Ph.D. from the University of California, Berkeley, and her B.S. from Tsinghua University. Her research interests include quantifying the central dogma, extracting hidden information from bulk, single-cell, and spatial multi-omics data, and ensuring statistical rigor in biomedical data analysis. She emphasizes using in silico negative controls to avoid false discoveries.
Jessica has received multiple awards in recognition of her work, including the NSF CAREER Award, Sloan Research Fellowship, Johnson & Johnson WiSTEM2D Math Scholar Award, MIT Technology Review 35 Innovators Under 35 China, Harvard Radcliffe Fellowship, COPSS Emerging Leader Award, and ISCB Overton Prize.
 Remo Rohs
Remo Rohs
USC
USA
Remo Rohs is a Professor of Quantitative and Computational Biology, Chemistry, Physics and Astronomy, and Computer Science at the University of Southern California. In his research, he integrates the fields of genomics and structural biology through studies of transcription factor binding to the genome. He and his team develop and apply methods that provide mechanistic insights into protein-DNA binding across multiple scales. These include the high-throughput prediction of DNA structural properties, statistical machine learning and deep learning methods to identity structural readout modes, molecular simulations, and experimental protein-DNA binding assays.
Rohs is also the Founding Chair of the Department of Quantitative and Computational Biology at the University of Southern California, where he leads the historically first Ph.D. Program in Computational Biology and Bioinformatics and a new undergraduate major in Quantitative Biology that he established. Rohs’ research is funded by the National Institutes of Health and the Human Frontier Science Program. Rohs is an Alfred P. Sloan Research Fellow and an elected Fellow of the American Association for the Advancement of Science.
 Paul Spellman
Paul Spellman
UCLA
USA
Dr. Spellman works to apply genomic and computational technologies to improve human health with a primary emphasis on improving outcomes for people with cancer. The work in his lab works at all phases from technology/method development, to application of technologies to answer critical questions in cancer biology, to population studies to understand the impact of genetic variation of disease, and to implementation trials that directly impact health. Current funded research focuses on systematic analysis of genetic and gene regulation information in clinical cohorts as part of the Genome Data Analysis Network and a clinical trial implementing genetic health screening for hereditary breast and ovarian cancer and Lynch syndromes. Other areas of interest include polygenic risk implementation and modeling, precision medicine, and understanding the molecular biology of cellular replication.
 David Van Valen
David Van Valen
Caltech
USA
Our group studies how living systems and their respective viruses encode and decode information about their internal state and their environment by combining ideas from cell biology and physics with recent advances in imaging, machine learning, and genomics to make novel measurements. Active research directions in our group include:
Host-virus interactions during viral decision making
How do viruses access information about their host cell's environment and internal state? How does this information flow to key decision points in the viral lifecycle? We are answering this question for temperate bacteriophage using functional genomics and high-throughput single-cell imaging.
Deep learning for single-cell biology
One of the major computational challenges of analyzing modern imaging experiments is image segmentation, that is determining which parts of a microscope image correspond to which individual cell. Our prior work has demonstrated that deep learning is a natural solution for this problem. We are currently developing the next generation of deep learning algorithms that can analyze dynamic data from live-cell imaging experiments as well as multi-dimensional data from spatial genomics experiments.
Integrated measurements of signaling and gene expression
Mammalian cells use dynamics to expand the information encoding capacity of their signaling networks, but how these dynamics are decoded into patterns of gene expression is less clear. Recent technological advances have made it possible to measure signaling dynamics and genome wide gene expression profiles in the same individual cell. We are working to merge live-cell imaging and spatial genomics data to quantify information transmission in signaling networks involved in the anti-viral response.