Conference on Semantics in Healthcare and Life Sciences (CSHALS)
Tutorial (hands on)
Updated February 24, 2012Special Offer: CSHALS 2012 Tutorial Registration
Wednesday, February 22
8:30 a.m. - 5:00 p.m.
Registration is now open for the February 22nd CSHALS Tutorial without the requirement of attending the main conference on February 23-24. If you decide after attending the Tutorial to also register for the main conference, 50% of your $250 Tutorial fee will be applied toward your on-site conference registration fee. This special Tutorial-only registration offer must be completed by Monday, February 20 (5:00 pm Eastern). Full details available at www.iscb.org/cshals2012-registration
Wednesday, Feb 22, 2012
8:30 am. - 5:00 p.m.
Location:
Charles Suite - 2nd Floor
Royal Sonesta Hotel
Semantic Healthcare and Life Sciences Tutorial: Mashing HC and LS Data
Hands-on Tutorial Synopsis
Presented by RPI
CSHALS Tutorial Coordinator: Lee Feigenbaum, Cambridge Semantics
RPI Tutorial Coordinator: Joanne Luciano, This email address is being protected from spambots. You need JavaScript enabled to view it.
Presenters:
- Dominic DiFranzo, This email address is being protected from spambots. You need JavaScript enabled to view it.
 Presentation (.pdf)
Presentation (.pdf)
- Jim McCusker, This email address is being protected from spambots. You need JavaScript enabled to view it.
- Joshua Shinavier. This email address is being protected from spambots. You need JavaScript enabled to view it.
 Presentation resource: http://services.fortytwo.net/linked-data
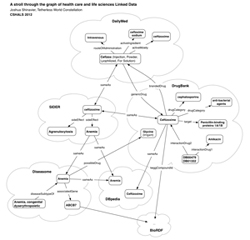
Presentation resource: http://services.fortytwo.net/linked-data Linked Data diagram (click here for PDF)
Linked Data diagram (click here for PDF)
Notice to Tutorial Participants:
In the tutorial, you will download and use an existing virtual machine (VM) that has been setup with an RDF store and other tools you will need. Before the tutorial, please install the Virtual Box software on your laptop. Virtual Box can be installed on Windows, Mac, or Linux laptops. To install Virtual Box, please follow the instructions at:
The tutorial will feature three parts:
- In part 1, you will be introduced to Semantic Web technologies, including RDF, RDFS, OWL, and SPARQL. You'll also learn how to issue basic SPARQL queries from JavaScript.
- In part 2, you will learn about Linked Data--Semantic Web data on the Web. This section will cover crawling and querying Linked Data, and then will take a look at health-related linked data such as DrugBank, DailyMed, SIDER, Diseasome, DBpedia, poss. LinkedCT, and PubMed. You'll also look at mashing up linked data with personal health records and talk about some use cases and demo applications.
- In part 3, you will build on what you've learned to accomplish these tasks:
- Convert a MAGE-TAB file from ArrayExpress to linked data and publish it using LODspeakr
- Add a summary graph to the experiment page to display the distribution of biospecimens by characteristic
- Load the data into GenePattern and extract differentially expressed genes
- Convert the gene list to RDF data cube and load into the triple store
- Create a heatmap from the gene list
[TOP]