links within this page: Kate Adamala | Aly Azeem Khan | Xihong Lin | Layla Oesper | Jian Zhou
 Kate Adamala
Kate Adamala
University of Minnesota
USA
Dr. Kate Adamala is a synthetic biologist and a McKnight Land-Grant Assistant Professor in the Department of Genetics, Cell Biology and Development at the University of Minnesota. Her research interests include astrobiology, synthetic cell engineering and biocomputing. Adamala is a co-founder and steering group member of the international Build-a-Cell Initiative, which seeks to broaden the impact of synthetic cell engineering. Find more information on Adamala’s lab at: http://www.protobiology.org
Programming life that is not alive: biocomputing in synthetic cells
Building a state-of-art ecosystem is pivotal for accelerating biological and health science discoveries and translational science. Such an ecosystem involves several pillars, including technology development, data fairness, scalable and powerful statistical and ML methods and tools, interpretable data analysis, and trustworthy decision-making. Rapid advancements in generative ML have revolutionized data utilization and enabled machines to learn from data more effectively. Statistics, as the science of learning from data while accounting for uncertainty, plays a pivotal role in addressing complex real-world problems and facilitating trustworthy decision-making. Massive multi-modal data have been generated in genomics and health. In this talk, I will discuss the challenges and opportunities in building such an ecosystem that integrates statistics, generative ML, and genomics and health. I will illustrate key points using the analysis of large scale biobanks, whole genome sequencing data, and electronic health records, and demonstrate the power of scientific discovery using ML-generated synthetic data. This talk aims to stimulate proactive and thought-provoking discussions, foster interdisciplinary collaboration, and cultivate open-minded approaches to advance scientific discovery.
 Aly Azeem Khan
Aly Azeem Khan
University of Chicago
USA
Aly A. Khan is an assistant professor at the University of Chicago, with appointments in the Departments of Pathology, Family Medicine, and the college. He develops computational methods to uncover clinically relevant insights about the immune system, translating these into improved diagnostic and therapeutic opportunities for human health. Previously, he was a research faculty member at the Toyota Technological Institute in Chicago, where he established a research program in computational immunology. In addition to his academic research, he has advanced translational science and research at Merck, Genentech, Tempus Labs, and currently 23andMe. Khan earned his Ph.D. in Computational Biology jointly from Cornell University and the Memorial Sloan Kettering Cancer Center. His contributions to computational immunology have been recognized with several honors, notably the National Institutes of Health DP2 NIAID New Innovator Award.
 Xihong Lin
Xihong Lin
Harvard University
USA
Xihong Lin is Professor and Former Chair of the Department of Biostatistics, Coordinating Director of the Program in Quantitative Genomics of Harvard TH Chan School of Public Health, and Professor of Statistics at Harvard University, and Associate Member of the Broad Institute of MIT and Harvard. Dr. Lin’s research interests lie in development and application of scalable statistical and computational methods for analysis of massive data from genome, exposome and phenome, such as large scale Whole Genome Sequencing studies, integrative analysis of different types of data, biobanks, as well as analysis of complex epidemiological and observational studies, and statistical learning methods for big data. She is an elected member of the US National Academy of Medicine. Dr. Lin received the 2002 Mortimer Spiegelman Award from the American Public Health Association, the 2006 Presidents’ Award and the 2017 FN David Award from the Committee of Presidents of Statistical Societies (COPSS), and an elected fellow of the American Statistical Association and the Institute of Mathematical Statistics. She is the recipient of the MERIT Award (R37) (2007-2015) and the Outstanding Investigator Award (R35) (2015-2022) from the National Cancer Institute, and the contact PI of the Harvard Analysis Center of the Genome Sequencing Program of the National Human Genome Research Institute. She has been active in COVID-19 research. She chairs the COPSS-National Institute of Statistical Science (NISS) COVID-19 Data Science Webinar series, jointly organized by all the leading North America Statistical Societies. Dr. Lin is the former Chair of the COPSS (2010-2012). She is the founding chair of the US Biostatistics Department Chair Group, and the founding co-chair of the Young Researcher Workshop of East-North American Region (ENAR) of International Biometric Society. She co-launched the Section of Statistical Genetics and Genomics of the American Statistical Association (ASA). She is the former Coordinating Editor of Biometrics, and the founding co-editor of Statistics in Biosciences. She is the former member of the Committee of Applied and Theoretical Statistics of the US National Academy of Science.
Accelerate Discovery by Integrating AI, Statistics and Genomic Health Science
Building a state-of-art ecosystem is pivotal for accelerating biological and health science discoveries and translational science. Such an ecosystem involves several pillars, including technology development, data fairness, scalable and powerful statistical and ML methods and tools, interpretable data analysis, and trustworthy decision-making. Rapid advancements in generative ML have revolutionized data utilization and enabled machines to learn from data more effectively. Statistics, as the science of learning from data while accounting for uncertainty, plays a pivotal role in addressing complex real-world problems and facilitating trustworthy decision-making. Massive multi-modal data have been generated in genomics and health. In this talk, I will discuss the challenges and opportunities in building such an ecosystem that integrates statistics, generative ML, and genomics and health. I will illustrate key points using the analysis of large scale biobanks, whole genome sequencing data, and electronic health records, and demonstrate the power of scientific discovery using ML-generated synthetic data. This talk aims to stimulate proactive and thought-provoking discussions, foster interdisciplinary collaboration, and cultivate open-minded approaches to advance scientific discovery.
 Layla Oesper
Layla Oesper
Carleton College
USA
Layla Oesper is an Associate Professor of Computer Science at Carleton College in Northfield, MN. Dr. Oesper received her B.A. in mathematics from Pomona College and her Sc.M and Ph.D. in Computer Science from Brown University. Dr. Oesper is also the recipient of NSF CRII and CAREER Awards. Her lab focuses on the design of computational methods related to inference and analysis of cancer evolution.
Computational methods for improved inference of tumor evolution
Tumors evolve as part of an evolutionary process where distinct sets of genomic mutations accumulate in different cell lineages descending from an original founder cell. A better understanding of how such tumor lineages evolve over time, which mutations occur together or separately, and in what order these mutations were gained may yield important insight into cancer and how to treat it. Thus, in recent years there has been an increased interest in computationally inferring the evolutionary history of a tumor – that is, a rooted tree where vertices represent populations of cells that have a unique complement of somatic mutations and edges that represent ancestral relationships between these populations. However, accurately inferring these trees from sequencing data is often a challenging process. In this talk, I will describe computational methods that address issues related to the inference and analysis of tumor evolution.
 Jian Zhou
Jian Zhou
UT Southwestern Medical Center
USA
Jian Zhou is an Assistant Professor in the Lyda Hill Department of Bioinformatics. He is a Lupe Murchison Foundation Scholar in Medical Research and is a Scholar of the Cancer Prevention and Research Institute of Texas (CPRIT). Prior to joining UTSW, he was a Flatiron Research Fellow at the Center of Computational Biology at Flatiron Institute, New York. He received his B.S. in Biological Sciences from Peking University and Ph.D. in Quantitative and Computational Biology from Princeton University. The Zhou lab works at the intersection of machine learning and genomics. The lab develops computational methods to improve our understanding of genome-based gene regulation and the genetic basis of human diseases. Currently the lab focuses on developing computational methods for improving predictive models for genomic sequence, understanding underlying biological mechanisms, and designing genomic sequences with desired properties. Advancing machine learning and AI methods for science is a long-term goal of the lab. Visit the lab website for more details https://zhoulab.io
Sequence-basis of transcription initiation in the human genome
Transcription initiation is a process that is essential to ensuring the proper function of any gene, yet we still lack a unified understanding of sequence patterns and rules that explain most transcription start sites in the human genome. By predicting transcription initiation at base-pair resolution from sequences with a deep learning–inspired explainable model called Puffin, we show that a small set of simple rules can explain transcription initiation at most human promoters. We identify key sequence patterns that contribute to human promoter activity, each activating transcription with distinct position-specific effects. Furthermore, we explain the sequence basis of bidirectional transcription at promoters, identify the links between promoter sequence and gene expression variation across cell types, and explore the conservation of sequence determinants of transcription initiation across mammalian species.
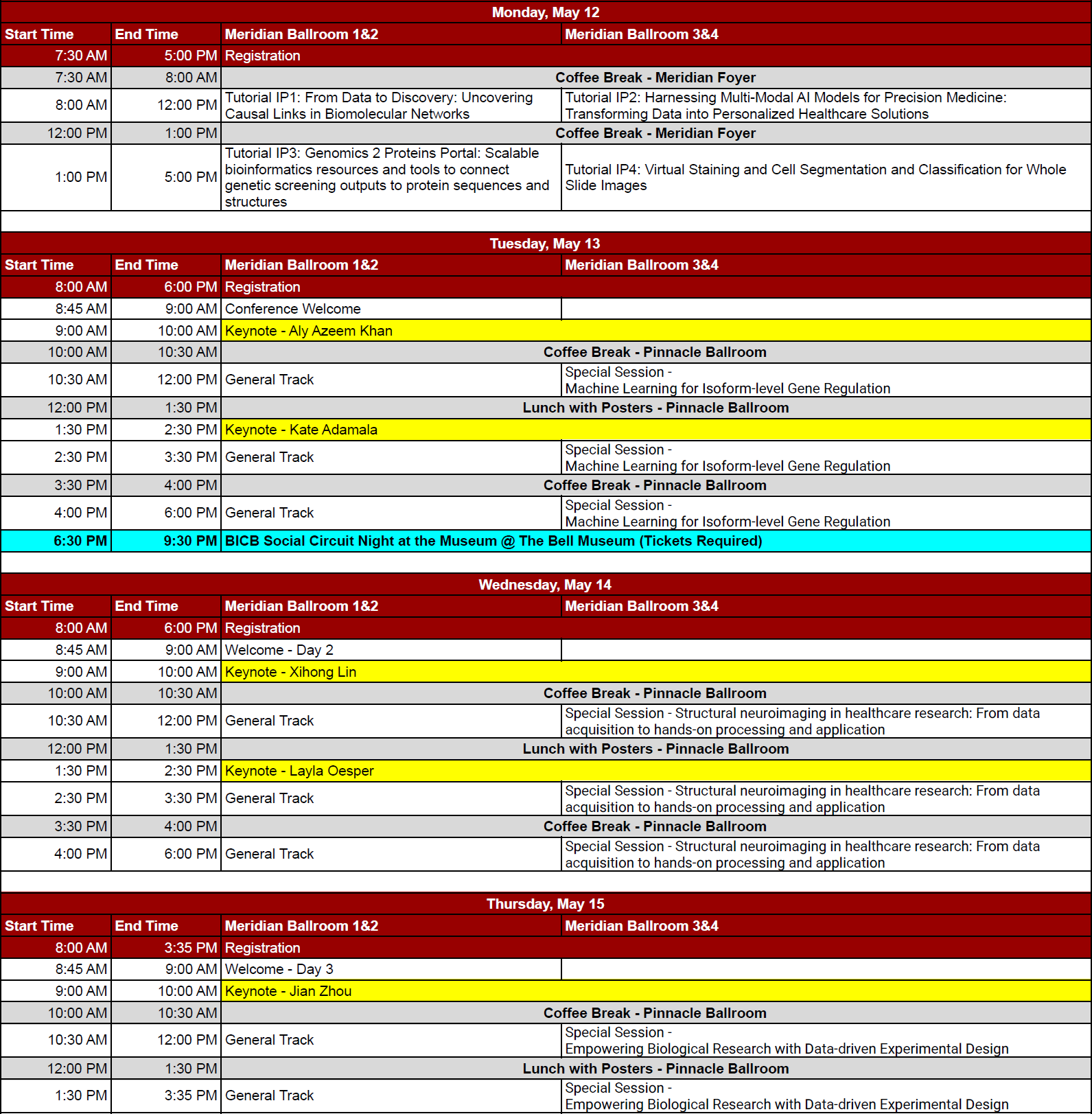
There will be a series of in-person and virtual tutorials prior to the start of the conference. Tutorial registration fees are shown at: https://www.iscb.org/ismbeccb2025/register#tutorials
In-person Tutorials (All times GMT)
- Tutorial IP1: From Data to Discovery: Uncovering Causal Links in Biomolecular Networks
- Tutorial IP2: Harnessing Multi-Modal AI Models for Precision Medicine: Transforming Data into Personalized Healthcare Solutions
- Tutorial IP3: Genomics 2 Proteins Portal: Scalable bioinformatics resources and tools to connect genetic screening outputs to protein sequences and structures
- Tutorial IP4: Virtual Staining and Cell Segmentation and Classification for Whole Slide Images
Room: Meridian Ballroom 1&2, Second Floor
Date: May 12, 2025 at 08:00
Organizer:
Karen Sachs
Max Participants: 30
Description
This workshop on causal discovery in biomolecular networks is designed to introduce participants to the principles and methodologies for uncovering causal relationships in biological regulatory networks, from molecular datasets. As the field increasingly relies on large-scale data from genomic, transcriptomic, and proteomic studies, the ability to decipher causal, mechanistic information from these datasets is essential for expanding our understanding of complex biological systems, and for maximally extracting information from multimodal high dimensional datasets.
This workshop will cover foundational concepts of causal modeling, with a focus on causal discovery: inference of the mechanism or structure of the regulatory networks which gave rise to the omics data observed. Participants will gain a clear and firm grasp of the fundamental principles of causal inference, learn about a variety of methods being used in the field, become aware of potential pitfalls and concerns with the techniques, and gain hands-on experience with real datasets. We will discuss practical methods to incorporate literature derived databases such as INDRA and OMNIPATH, to integrate information from these prior knowledge networks into analyses of molecular datasets. In the practice session, we will apply causal discovery methods to elucidate mechanisms underlying biological processes.
Prerequisites
For the practice session, some fundamental knowledge of programming is required.
Attendees can join without participating in the practice session.
For the theoretical portion, some familiarity with probability is helpful but not necessary.
Materials
Attendees will be provided with optional reading materials. Each participant for the practice session is encouraged to bring their own data for the practice session.
Tutorial IP2: Harnessing Multi-Modal AI Models for Precision Medicine: Transforming Data into Personalized Healthcare Solutions
Room: Meridian Ballroom 3&4, Second Floor
Date: May 12, 2025 at 08:00
Organizer:
Danielle Stover
Max Participants: 100
Description
This tutorial will focus on the rapidly advancing field of multi-modal modeling for precision medicine, a critical area where artificial intelligence is reshaping the landscape of personalized healthcare. The session will delve into the integration of diverse data types—including genomics, imaging, electronic health records (EHRs), and patient-reported outcomes—to improve diagnosis, treatment, and prognosis in precision medicine.
The research theme centers around how to effectively use AI to combine these disparate data sources into a unified framework, providing deeper insights into patient-specific treatment responses and disease mechanisms. Attendees will learn about the latest advancements in machine learning models (e.g., transformers, graph neural networks) tailored for multimodal data fusion, as well as practical strategies for implementing these models in real-world medical settings.
The session is designed for participants including graduate students, postdoctoral researchers, bioinformatics professionals, clinical data scientists, and industry practitioners. It is especially suitable for those with a foundational understanding of machine learning and a keen interest in advancing their knowledge of AI applications in healthcare.
The tutorial’s timeliness lies in the ongoing explosion of healthcare data and the need for sophisticated AI approaches to manage and leverage this data effectively. The bioinformatics community stands at the forefront of this effort, and the tutorial aims to bridge the gap between cutting-edge AI techniques and their real-world application in precision medicine. This session will empower participants to develop models that can drive more personalized, effective, and efficient patient care.
Prerequisites
A preliminary understanding of python and machine learning
Materials
Attendees are encouraged to plan to bring their laptop if they choose to participate in coding sessions
Tutorial IP3: Genomics 2 Proteins Portal: Scalable bioinformatics resources and tools to connect genetic screening outputs to protein sequences and structures
Room: Meridian Ballroom 1&2, Second Floor
Date: May 12, 2025 at 13:00
Organizer:
Sumaiya Iqbal
Max Participants: 40
Description
This tutorial introduces the Genomics 2 Proteins (G2P) portal, a bioinformatics tool that connects genetic variants with protein sequences and structures, enabling researchers to interpret genomic data in the context of protein function.
Participants will learn to integrate multi-omics datasets – genomics, proteomics, and structural biology – to investigate the structural implications of genetic mutations.
Hands-on sessions will guide participants in navigating G2P modules, mapping genetic variants to structural changes in proteins, visualizing protein features, and using APIs for advanced applications.
Case studies will showcase real-world applications, such as mapping clinical mutations and interpreting base-edited variants.
Prerequisites
Sessions I-III: No prerequisites are required, though familiarity with bioinformatics databases like UniProt, AlphaFoldDB, and structure visualization tools (e.g., PyMOL) is beneficial.
Session IV (Advanced Coding): Basic knowledge of Python or bash/shell scripting is recommended for participants interested in the coding and API components.
Materials
Attendees are expected to bring their own laptops to participate in interactive exercises and follow along with case studies
Room: Meridian Ballroom 3&4, Second Floor
Date: May 12, 2025 at 13:00
Organizer:
Quincy Gu
Max Participants: 100
Description
Artificial Intelligence (AI) has profoundly impacted the field of digital pathology. One innovative AI-driven technology, virtual staining, can convert unstained tissues into stained ones or change one type of stain to another. In clinical diagnostics, hematoxylin and eosin (H&E) stained whole slide images (WSIs) serve as the gold standard for cancer diagnosis, often followed by special staining WSIs, such as immunohistochemistry (IHC) or multiplexed WSIs, to assess patients' genetic mutation status and aid in selecting personalized treatment options. However, the special staining process is time-consuming and requires adjacent tissue samples, making it challenging to directly apply AI models developed for cell segmentation and classification on H&E-stained WSIs to special stained WSIs. Virtual staining, which can digitally convert H&E stained WSIs into other specially stained formats (like IHC or multiplexing), provides an ideal solution for cell segmentation and classification tasks without needing physically stained samples, enhancing single-cell analysis. Due to time constraints and resource limitations, this tutorial will focus solely on deep learning-based methods for cell segmentation and single-cell analysis on multiplexed WSIs.
Prerequisites
The interactive hands-on tutorial will require the internet access for the audience
The audience are highly recommended to bring their laptop to practice in the session with the instructor’s assistance
Material
Python 3.10 version or later installed on their laptop if they prefer to have a more interactive hands-on session with the instructors.
Conference Steering Committee:
- Bruce Aronow, Children's Hospital Medical Center, University of Cincinnati
- Gary Bader, University of Toronto
- Robert Blumenthal, University of Toledo
- Serdar Bozdag, University of North Texas
- Sarath Chandra Janga, Indiana University Purdue University
- Maria Chikina, University of Pittsburgh
- Sorin Draghici, Wayne State University
- Dannie Durand, Carnegie Mellon University
- Bruno Gaeta, University of New South Wales (Ex Officio)
- Anthony Gitter, University of Wisconsin-Madison
- Daisuke Kihara, Purdue University
- Dan Knights, University of Minnesota
- Jundong Liu, Ohio University
- Shaun Mahony, Pennsylvania State University
- Tijana Milenkovic, University of Notre Dame
- Chad Myers, University of Minnesota
- Layla Oesper, Carleton College
- Aïda Ouangraoua, Université de Sherbrooke
- John Parkinson, Hospital for Sick Children
- Catherine Putonti, Loyola University
- Predrag Radivojac, Northeastern University
- Sushmita Roy, University of Wisconsin-Madison
- Russell Schwartz, Carnegie Mellon University (Co-chair)
- Lonnie Welch, Ohio University (Co-chair)
- Jeff Xia, McGill University
- Feng Yue, Northwestern University
Program Committee:
- Coming Soon!
- Chad Myers, University of Minnesota
- Feng Yue, Northwestern University
Publication Chair:
- Mingfu Shao, Penn State University
Poster Chairs:
- Guenter Tusch, Grand Valley State University
- Helen Piontkivska, Kent State University
Travel Fellowship Chairs:
- Dan DeBlasio, Carnegie Mellon University
- Getiria Onsongo, Macalester College