Complete List of Winners - ISCB Art in Science Competition
| 2014 FIRST PLACE |
Hai Fang
Julian Gough
University of Bristol, United Kingdom
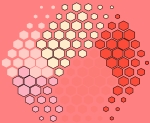
supraHex
This artwork called ‘supraHex’ is inspired by the prevalence of natural objects such as a honeycomb or at Giant’s Causeway. supraHex has architectural design of a supra-hexagonal map: symmetric beauty around the center, from which smaller hexagons radiate circularly outwards. In addition to this architectural layout, supraHex also captures mechanistic nature of these objects: formation in a self-organising manner. For this, supraHex is able to self-organise the input data (eg transcriptome data). In doing so, genes with similar data patterns are clustered to the same or nearby nodes (hexagons). The map distance (the hexagon size) tells how far each node is away from its neighbors, thus characterising relationships between clustered genes. Based on this map distance, supraHex is also able to partition the map to obtain gene meta-clusters covering continuous regions, as colour-coded by the ‘potato-peach-tomato’ colormap. This artwork is generated by an open-source R/Bioconductor package ‘supraHex’ (http://bioconductor.org/packages/release/bioc/html/supraHex.html).

| 2015 FIRST PLACE |
Luke Wilson
Jim Procter
Geoff Barton
University of Dundee, United Kingdom
Analogue Alignment
“Multiple sequence alignments were once performed manually, and even today, we still examine automatically computed alignments to check that we can't do better.” –Jim Procter This is an image of the Jalview Abacus, a sculptural attempt to visually represent the function of the Jalview protein alignment program. The program can be used to find alignments of amino acids in similar proteins. These alignments are then used to find similarities and differences between these proteins.
This object expresses the core process of Jalview in a physical space, and plays on the relationship between high tech and low tech solutions. It is a functioning abacus built by hand from wood and steel. Each row is an extract from different similar proteins (cysteine proteases) and an alignment can be found by lining up the beads of like amino acids in the columns. If it was long enough it could be used to align the entire sequence manually.
Photography: Luke Wilson
Design and Construction: Luke Wilson in collaboration w. Jim Procter and Geoff Barton
| 2016 FIRST PLACE |
 |
Sean O'Donoghue, CSIRO & Garvan Institute, Australia
Christopher Hammang, Garvan Institute, Australia
Julian Heinrich, CSIRO, Australia
The Dark Proteome
Here, we use light and darkness to represent the known and unknown proteome of structural biology. Currently, only 12% of the human proteome has been observed with experimental structure determination methods such as crystallography or NMR spectroscopy. For a further 36%, structural information can be inferred by homology modelling. The remaining 52% of the proteome is 'dark', i.e., has completely unknown molecular conformation.
| 2017 FIRST PLACE |

Nick Schurch
Chris Cole
University of Dundee, United Kingdom
ImpactFactor
impactFactor is a perspective on the use of the Journal Impact Factor scores in science. In taking a literal interpretation of this score we, as scientists, are questioning its use as a measure of scientific quality or importance.
Journal Impact Factor aims to reflect the importance of a journal, however it is now used to assess the quality of the scientists who publish within it. Employers and funding bodies conflate this artificial metric as a simplistic judge of a scientist’s quality with their choice of publisher. With this perspective for modern scientists, it could be argued that where they publish has become more important than the science itself! We feel it would be much better to judge both the scientist and their publications on merit alone.
The use of artificial quality metrics is an issue for all scientists and this piece is part of the discussion of moving scientific publishing away from its 17th Century roots. Publication remains a cornerstone of scientific research but impactFactor highlights the need for a better way to judge, and, perhaps publish, impactful science.
The original acrylic on canvas artwork was exhibited at Symbiosis, a local collaborative Science/Art exhibition in Dundee. The piece measures 2m x 2m and is too big to bring to the conference, instead, we present here a 70cm x70cm photographic interpretation of the piece.

| 2018 FIRST PLACE |
Ruth Callaway
Swansea University, Biosciences, UK
Mondrian’s Sum of Squares
Science inspired many artist, but here it was the other way around. The visualisation of marine biodiversity data was modelled on paintings by the early 20th century Dutch artist Piet Mondrian. It is an ongoing challenge for ecologists to compress and simplify complex data and to illustrate patterns in marine ecosystems. Differently coloured and sized rectangles and squares were assembled in this Mondrian’s Sum of Squares and simultaneously shows numerical and taxonomic information of a benthic invertebrate seafloor community (Swansea Bay, Wales, UK). Each field, large or small, represents a different species. The size of the square or rectangle indicates how numerically common a species was, and colours indicate taxonomic or functional groups (blue: polychaete worms, yellow: bivalves, red: crustaceans, white: other mobile species, grey: other sessile species). The few large squares highlight that the seafloor community consists of just a handful of common species, while most occur in low densities. The overwhelming number of blue fields shows the importance of worm species for biodiversity. Like many of Mondrian’s paintings, this artwork is an abstract representation of the natural world. It differs in that Piet Mondrian deliberately stepped away from reality, while this work translates scientific data into art.
 |
| 2018 SECOND PLACE |
Alaa Abi Haidar
University of Pierre and Marie Curie
dEYEversity
The two contrasted eyes are composed of the same ingredients and diversity of eyes, ad infinitum. The artist owns all images’ rights.
Featured at La Nuit de la Photographie Contemporaine and soon in a gallery.
As for the technique, I developed image processing algorithms to crop the eyes from the 1001faces.org project to have them automatically reassembled in this mosaic using another algorithm that optimizes the images' position according to the best matching pixel intensities.
| 2018 THIRD PLACE |
Marwan Abdellah
École polytechnique fédérale de Lausanne (EPFL)
In Silico Brainbow
In silico brainbow optical section of a neocortical slice (920 × 640 × 1740 μm3) created with a virtual light-sheet fluorescence microscope (LSFM).
The simulation of the LSFM is performed on a physically-plausible basis using Monte Carlo ray tracing and geometric optics. The tissue model is reconstructed in a three-step process: 1) converting the morphological skeletons of the neurons into piecewise surface meshes that represent their membranes, 2) reconstructing a volumetric model of the tissue using solid voxelization and finally 3) tagging the neurons with the optical properties of the neocortical tissue and also the spectroscopic properties of different fluorescent dyes.
The slice is virtually-tagged with six different fluorescent proteins (GFP, CFP, eCFP, mBanana, mCherry and mPlum) and illuminated at the maximum excitation wavelength of each respective dye
| 2019 FIRST PLACE |
 |
Dr. Kliment Olechnovic
Department of Bioinformatics, Life Sciences Center
Vilnius University
Lithuania
Disassembled Tessellation