DREAM PRESENTATIONS & ABSTRACTS
Presented on Monday, November 10 and Tuesday, November 11
Updated Oct 28, 2014
--> Go directly to Tuesday, Nov 11
| MONDAY, NOVEMBER 10 |
9:00 am – 9:20 am
DR T01
The DREAM Rheumatoid Arthritis Responder Challenge: motivation, data, scoring, and results
Lara Mangravite1
1Sage Bionetworks
Rheumatoid Arthritis (RA) is a debilitating autoimmune disease that manifests through proinflammatory joint damage and for which reduction in inflammaRheumatoid Arthritis (RA) is a debilitating autoimmune disease that manifests through proinflammatory joint damage and for which reduction in inflammation is essential to prevent long-term deleterious effects. Standard of care treatment includes a class of drugs that block the inflammatory cytokine tumor necrosis factor-a (anti-TNF therapies) but nearly a third of patients fail to respond to these therapies. While it is known that patients with more severe disease tend to exhibit stronger response, there is not sufficient information available to develop prognostic biomarkers capable of predicting response a priori. Recent analyses indicated that ~25% of the variation in anti-TNF response in RA patients is heritable, suggesting that genetic factors may be useful in predicting treatment response. The DREAM RA Responder Challenge was designed to use a crowd-sourced modeling approach to assess whether common genetic variation could be used to improve on clinical prediction of response to anti-TNF therapy. In the Team Phase (February – June 2014), self-aggregated teams competed to build models that predicted (a) change in disease activity score (DAS28) from baseline following anti-TNF treatment or (b) responder status as determined according to EULAR criteria for end and delta DAS. Participants trained models using genetic and clinical data collected from a cohort of 2031 anti-TNF treated RA patients. Predictions were then tested using a second dataset collected from 723 patients that participated in the Consortium of Rheumatology Researchers of North America (CORRONA) CERTAIN study. The use of Gaussian process regression resulted in the most accurate predictions for change in DAS (r=0.39) and for responder status (ROC-AUC = 0.62) in the CORRONA study. Comparison of these results to a simple clinical model that used baseline DAS to predict change in DAS (r=0.35) suggests that the genetic contribution to the predictive power of these models was not large. In the Community Phase (July – October 2014), the eight teams with the most predictive models joined together in collaboration to directly assess the genetic contribution to predictions of treatment response. The results of this phase will be revealed at the DREAM conference.
...............................................................................................................................
Monday, November 10
9:20 am – 9:30 am
Awards Rheumatoid Arthritis Challenge
Stephen Friend
...............................................................................................................................
Monday, November 10
9:30 am – 9:50 am
DR T02
DREAM Best Performer Talk – Rheumatoid Arthritis Challenge
A generic method for predicting clinical outcomes and drug response
Fan Zhu1, Yuanfang Guan1
1University of Michigan
We developed an elegant Gaussian Process Regression (GPR)-based model to predict clinical outcomes and drug response. We applied it in both the genetics-only task and the genetics + clinical information-combined task in the RA challenge. It achieved the top accuracy in both the leaderboard and the final previously unseen test set, for both delta DAS and non-responder predictions. We will discuss the properties of this method, its application, and the evaluation results of GPR and several related methods.
...............................................................................................................................
Monday, November 10
9:50 am – 10:10 am
DR T03
DREAM Best Performer Talk – Rheumatoid Arthritis Challenge Session
Predicting response to Arthritis treatments: regression-based Gaussian processes on small sets of SNPs
Javier García-García1
1Universitat Pompeu Fabra, Barcelona, Catalonia, Spain
The aim of our study was to identify candidate SNPs playing a role in the response to therapy in rheumatoid arthritis (RA) patients, by compiling several sources of information such as the localization in the coding/non-coding region of the gene and its consequences in the translated protein (i.e., a synonymous or non-synonymous mutation). Genes affected by SNPs were first analyzed in order to select the most relevant associations with RA as follows. An initial list of potential candidates was selected using association analysis derived from the experimental data provided by the DREAM challenge. Additionally, we used multiple external sources of biomedical data to filter candidate SNPs. The list of candidates was expanded using gene priorization algorithms that combined protein-protein interaction networks and expression data. The procedure is based on the guilt-by-association principle and we selected from the extended list only those candidates with known SNPs. After the selection of genes, we used all SNPs reported for these genes. The resulting SNPs, in combination with clinical data, were used to predict the patients' response to treatments by means of regression-based models and a 10-fold cross-validation on the training dataset provided by the DREAM challenge. When models were applied to an independent dataset (the leaderboard set), their predictive power decreased significantly, pointing out a problem of overfitting in the model. After comparison of the initial list of potential candidates and the use of external sources of information (i.e., biomedical data to filter the candidate list and extending the list using guilt-by-association principles), we confirmed that the predictive value of the original list of candidate SNPs was not improved by any of the external information. Therefore, we simplified the approach and reduced the SNP list by selecting only those showing the highest Pearson's correlation with the patients' response (ÄDAS) in the leaderboard set (only about 20% of the initial SNPs). In the final independent dataset (CORRONA dataset) we achieved an AUC-ROC value of 0.6237 and AUC-PR value of 0.5071.
...............................................................................................................................
Monday, November 10
10:10 am – 10:20 am
Rheumatoid Arthritis Challenge Discussion
...............................................................................................................................
Monday, November 10
11:30 am – 11:50 am
DR T04
The ICGC-TCGA DREAM Somatic Mutation Calling Challenge: motivation, data, scoring, and results
Paul Boutros1,2
1Ontario Institute for Cancer Research, 2University of Toronto
The detection of somatic mutations from cancer genome sequences is a major bottleneck to the routine implementation of clinical-sequencing and to the discovery of mutations associated with patient survival and response to therapy. Benchmarking somatic mutation detection algorithms is complicated by the lack of gold-standards, extensive resource requirements and difficulties in sharing personal genomic information. To resolve these issues, we launched the ICGC-TCGA DREAM Somatic Mutation Calling Challenge — a crowd-sourced benchmark of somatic mutation detection algorithms. We report here the initial results from the Challenge, focusing on the first four tumors. We highlight the development of BAMSurgeon, the first tool for simulating cancer genomes. We find that ensembles of different analysis pipelines outperform even the best pipeline, that different algorithms exhibit characteristic error profiles, and that false positives can be biased in ways that significant confound discovery of specific biological mutation signatures known to be found in human tumors. We also give a status update on the new community phases to the Challenge that will be launching in 2015.
...............................................................................................................................
Monday, November 10
12:00 Noon – 12:20 pm
DR T05
DREAM Best Performer Talk – Somatic Mutation Calling Challenge
novoBreak: robust characterization of structural breakpoints in cancer genomes
Zechen Chong1, Ken Chen1
1University of Texas MD Anderson Cancer Center
Structural variation (SV) is a major source of genomic variation and plays a driving role in cancer genome evolution. However, the current strategy of using next-generation whole genome sequencing still does not achieve the comprehensiveness and sensitivity required to identify abundant SV breakpoints in heterogeneous tumor samples. This is due to challenges in acquiring high sequencing depth as well as methodological limitations in aligning and interpreting short reads spanning breakpoints. To alleviate such challenges and to deepen our understanding of cancer genome evolution, we developed a novel algorithm, novoBreak, which targets the reads that substantially differ from the normal genome reference and outputs the “breakome”: the collection of genomic sequences spanning breakpoints and unobserved in the reference alignment. novoBreak can comprehensively characterize a variety of breakpoints that are introduced by small indels, large deletions, duplications, inversions, insertions, and translocations at base-pair resolution from whole genome sequencing data. In contrast to most existing SV discovery programs such as Delly and Meerkat, novoBreak first clusters reads around potential breakpoints and then locally assembles the reads associated with each breakpoint into contigs. After aligning the contigs to the reference, novoBreak then identifies the precise breakpoints and infers the types of SVs. novoBreak performs substantively better than other widely used algorithms and ranked at No. 1 in the recent ICGC-TCGA DREAM Somatic Mutation Calling Challenge. The higher sensitivity of novoBreak makes it possible to uncover a large number of novel and rare SVs, as shown in our data from The Tumor Genome Atlas (TCGA) and from the 1000 Genomes project. Wider application of novoBreak is under way and is expected to definitively reveal the comprehensive structural landscape that can be linked to novel mechanistic signatures in cancer genomes
...............................................................................................................................
Monday, November 10
12:20 pm – 12:40 pm
DR T06
DREAM Best Performer Talk – Somatic Mutation Calling Challenge
Application of MuTect for sensitive and specific somatic point mutation detection in DREAM challenge synthetic data
Mara Rosenberg1, Kristian Cibulskis1, Adam Kiezun1, Louis Bergelson1, Gad Getz1,2
1Broad Institute of Harvard and MIT, 7 Cambridge Center, Cambridge, MA 02142, USA
2Department of Pathology, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA
Sensitive and specific detection of somatic point substitutions is a critical aspect of characterizing the cancer genome. However, tumor heterogeneity, purity and sequencing errors confound the confident identification of events at low allelic fractions. MuTect, a previously described method for somatic mutation calling [1], allows for high sensitivity by first implementing a Bayesian classifier and then further reducing the false positives through carefully tuned filters. We applied MuTect to the four synthetic datasets in the DREAM challenge and achieved top scoring performance with specificity ranging from 0.98 to 0.99 and sensitivity from 0.74 to 0.97, consistent with our experience with real data. This had a corresponding false positive rate between 0.01 and 0.07 mutations per Mb. Here, we will describe our approach that used an application of MuTect and filters to reduce artifacts from bam alignment errors and base specific sequencing noise.
...............................................................................................................................
Monday, November 10
12:40 pm – 12:50 pm
Somatic Mutation Calling Challenge Discussion
...............................................................................................................................
Monday, November 10
3:00 pm – 3:20 pm
DR T07 - DREAM Challenge Introduction Talk – AML Challenge Session
The DREAM AML Outcome Prediction Challenge: Motivation, Data, Scoring and Results
Amina Ann Qutub1
1Rice University
In 2014, there will be 18,860 new cases of acute myeloid leukemia (AML), and 10,460 deaths from AML. There is urgency in finding better treatments for this type of leukemia, as only about a quarter of the patients diagnosed with AML survive beyond 5 years. The goal of the DREAM 9 Acute Myeloid Leukemia (AML) Outcome Prediction Challenge is to harness the power of crowd-sourcing to speed the pace of diagnosing and treating AML. Participants predict patient outcomes from a high-dimensional proteomics and clinical dataset for AML. Results of the Challenge include predictive clinical models that surpass current standards; new methods to handle high-dimensional clinical data; and insight into markers of AML and potential new cancer drug targets.
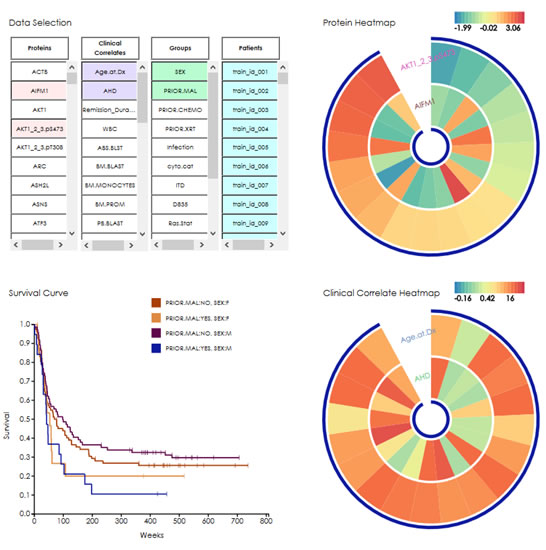 |
| Figure 1. Acute Myeloid Leukemia Outcome Prediction DREAM Challenge Data. Tools provide DREAM participants the high-dimensional leukemia dataset (191 patients, 40 clinical attributes, 230 protein expression levels) in an interactive format. Participants predict CR/PR, remission duration and overall survival (bottom left) as function of clinical correlates and/or RPPA data. |
DREAM 9 participants were provided a dataset of 190 AML patients seen at M.D. Anderson Cancer Center, and treated with ARA-C therapy (Fig. 1). The dataset includes 40 clinical correlates and the expression level of 231 proteins probed by RPPA protein array analysis. This AML dataset provides information that enables researchers for the first time to link protein signaling with mutation status and cytogenetic categories – offering DREAM Challenge participants the potential to surpass existing methods in identifying drug targets and tailoring therapies for cancer patient subpopulations. Challenge participants were posed three questions based on this data: to predict which AML patients will be primarily resistant to therapy and which patients will have complete remission; to predict remission duration; and to predict overall survival. Baseline predictive models of AML outcome were provided to participants. Each week, teams predicted outcomes for 100 representative patients whose outcome was withheld, based on their choice of clinical and proteomic features. These predictions were scored against the test data using two statistical comparisons for each Challenge question: balanced accuracy and AUROC for SubChallenge 1 and the concordance index and Pearson correlation coefficient for SubChallenges 2 and 3. The best performing algorithms were determined by the average rank over these two metrics and the sum of the normalized metrics.
Results of the top-scoring algorithms provide insight into the main factors determining AML outcome, both with and without proteomic data included. Notably, top performers predicting complete remission or relapse incorporated proteomics in their models. On the other hand, most participants only considered clinical correlates when predicting outcome over time (i.e., remission duration and overall survival). The ongoing extended phase of the Challenge rephrased the three SubChallenges into a single classification problem, with incentives for incorporating the RPPA data. Results of the AML Outcome Prediction Challenge also illuminated the performance of specific model types. Four baseline statistical models with no parameter optimization were provided to all participants. These consisted of logistic regression, Random Forest, decision tree with adaptive boosting and support vector machine. Median and mode imputation was used to replace missing patient data values. This talk will briefly introduce and show the performance of the diverse baseline and submitted models on the clinical data. It will also highlight which groups of models performed better on specific patient populations, and which patients’ outcomes were consistently predicted well. In sum, the Challenge provided new approaches to predict leukemia patients’ outcome and interpret RPPA data – and it has identified key clinical and proteomic markers of AML.
...............................................................................................................................
Monday, November 10
3:30 pm – 3:50 pm
DR T08
DREAM Best Performer Talk – AML Challenge
Evolution-informed modeling to predict AML outcomes
Li Liu1
1Arizona State University
As a part of the DREAM9 Challenge, the Acute Myeloid Leukemia (AML) Outcome Prediction Subchallenge 1 aims to foretell if an AML patient will have a complete response or resistance to treatment based on 40 clinical covariates and 231 proteomic measurements. Previous analysis performed by the challenge organizers showed that the high level of noise in proteomic data reduced their predictive power when used in an uninformed manner. To solve this problem, I designed an evolution-informed model that incorporates weights derived from evolutionary conservation and univariate analysis in machine learning algorithms.
Based on evolutionary patterns of cancer genes, it can be inferred that changes of expression levels may have more profound impact if they involve conserved proteins, as compared to variable proteins. Therefore, higher weights can be given to slow-evolving proteins, and to proteins differentially expressed between two outcome groups. To estimate protein conservation, evolutionary rate (r) for each position in each protein was calculated based on alignments of orthologous sequences from 46 vertebrates. The evolutionary weight (WE) of a protein was the reciprocal of average evolutionary rate over all positions ( ). Clinical variables took the maximum of all WEs. Next, Student’s t-test was performed for each feature. P-values were transformed via negative logarithm (-log(P)) and used as the differential weight (WD). For a given feature, the final weight was the sum of its evolutionary and differential weights (W = WE + WD).
In the feature selection step, each feature was first transformed to z scores, and then multiplied with its corresponding weight (W). Because the training data were highly unbalanced, an ensemble approach was used to construct multiple classification models with balanced subsamples. Using stability selection with sparse logistic regression, features identified in >50% of bootstrapping runs were selected. In the classification step, these features with un-weighed values were used to construct a random forest model with 50 trees. The above procedure was repeated 100 times to produce an ensemble of 100 random forest models. Given a patient, 100 predictions were obtained, one from each model. The confidence score equals the proportion of models that predict the patient to have a complete response.
When applied to test data that are blind to participants of this challenge, this evolution-informed model achieved a balanced accuracy of 77.9% and AUROC of 0.795, ranked number one among all participants. Features that were selected in more than 80% of all models include Chemo (Flu-HDAC), cyto.cat (21), CD34, cyto.cat (-5), Age.at.Dx, ABS.BLST, PIK3CA, and GSKA_B.pS21_9.
...............................................................................................................................
Monday, November 10
3:50 pm – 4:10 pm
DR T09
DREAM Best Performer Talk – AML Challenge
Acute myeloid leukemia outcome prediction via dictionary learning for sparse coding
Zhilin Yang1, Subarna Sinha2 and David L. Dill2
1Tsinghua University, 2Stanford University
This challenge was to use clinical and reverse-phase protein array (RPPA) data to solve three subchallenges: predicting complete remission after treatment, predicting remission duration, and predicting survival time. We describe our solutions to the first two subchallenges. For the first challenge, we found that a support vector machine (SVM) classifier with the radial basis function kernel was the most effective standard classifier of those we tried. We added a manual rule that any patient treated with Flu-HDAC would experience remission.
We found it difficult to improve prediction performance using RPPA data until we used dictionary learning for sparse coding, which learns low-rank latent state vectors from the original data in an unsupervised way and represents each sample as a sparse linear combination of the latent states, for feature extraction. Using sparse coding features of all protein data improved classifier performance. Applying sparse coding to pathway-specific subsets of proteins improved performance further, showing that prior knowledge of pathways can be useful in this task. Interestingly, some of the latent states in the pathway-specific sparse codes seemed to be biologically meaningful. The quality of the results also depended on a hybrid feature selection algorithm for clinical variables to avoid mixing up continuous and categorical features. We observed significant batch effect in the RPPA data, which we tried to correct unsuccessfully using several standard methods.
For the second subchallenge, we used an average of three support vector regressions using different subsets of the features. We were unable to improve the quality of predictions in this subchallenge using the RPPA data.
...............................................................................................................................
Monday, November 10
4:10 pm – 4:30 pm
DR T10
DREAM Best Performer Talk – AML Challenge
A bagged, semi-parametric model to predict survival time for acute myeloid leukemia patients
Xihui Lin1, Gregory M. Chen1, Honglei Xie1, Geoffrey A. M. Hunter1, Paul C. Boutros1,2
1Ontario Institute for Cancer Research, 2University of Toronto
While many AML patients go into remission after treatment, survival time remains highly variable across individuals. Predicting these differences would be of major clinical value in personalizing therapy. As part of the ninth Dialogue for Reverse Engineering Assessment and Methods (DREAM9) challenge, we sought to accurately estimate the survival of AML patients by integrating clinical and proteomic features. We initially formulated survival models based on random forests, boosted quantile regression, and weighted linear models, but these performed no better than a benchmark Cox model with only five clinical variables. Therefore, we decided to extend the benchmark Cox model for our final submission in the DREAM9 challenge. Specifically, we used a bootstrap aggregated (bagged) modified Cox model based on five clinical features: age at diagnosis, Anthra based treatment administered, hemoglobin count, Albumin levels, and cytogenic category. Researchers identified cytogenics as the single most important prognostic factor in AML patients; however, the cytogenic categories in the data for the challenge were imbalanced. To resolve this, we re-stratified patients into high, intermediate, intermediate-low, and low risk survival groups based on their cytogenic category. This significantly improved the predictive power of the model. Surprisingly, incorporating additional clinical and/or proteomic features in the Cox model diminished its performance. These results suggest that our reclassified cytogenic categories can improve predictions of patient survival and, hence, might be the key to help tailor therapies for AML patients.
...............................................................................................................................
Monday, November 10
4:30 pm – 4:40 pm
AML Challenge Discussion
...............................................................................................................................
Top of Page
| TUESDAY, NOVEMBER 11 |
9:30 am – 9:50 am
DR T11
The Broad-DREAM Gene Essentiality Prediction (GEP) Challenge: motivation, data, scoring and results
Mehmet Gönen1
1Oregon Health and Science University
The translation of cancer genomic data systematically into cancer therapies remains a challenge. Large-scale functional screening of cancer cell lines provides a complementary approach to cancer genome studies that aim to characterize the molecular alterations (mutations, copy number alterations, basal gene expression, etc.) of primary tumors. Project Achilles, one such functional screen, aims to link gene dependencies to the molecular characteristics of each cancer in order to identify molecular targets and guide therapeutic development. The promise of targeted cancer therapy requires both effective treatments and good biomarkers to identify patient populations likely to respond to those treatments. Therefore, a critical need exists to accurately predict essential genes across a wide variety of cancer subtypes.
The goal of this challenge is to use a crowd-based competition to develop predictive models that can infer gene dependency scores in cancer cells (i.e., genes that are essential to cancer cell viability when suppressed) using features of those cell lines. Participants were asked to solve three sub-challenges:
1. Build a model that best predicts the gene essentiality values of genes, using the molecular characteristics/features of the cancer cell lines.
2. Identify the most predictive features for each gene essentiality of a prioritized list of genes.
3. Identify the most predictive features for all gene essentiality values of a prioritized list of genes.
We had submissions from 21, 13, and 14 teams for sub-challenges 1, 2, and 3, respectively. Here, we present the evaluation methodology and results for all sub-challenges.
...............................................................................................................................
Tuesday, November 11
9:50 am – 10:00 am
Awards GEP Challenge
Bill Hahn
...............................................................................................................................
Tuesday, November 11
10:00 am – 10:20 am
DR T12
DREAM Best Performer Talk – GEP Challenge
Learning kernel-based feature representation for gene essentiality prediction
Masayuki Karasuyama1, Hiroshi Mamitsuka1
1Kyoto University, Japan
We develop a predictive method for estimating gene essentiality, focusing on learning a predictive feature representation. Our method uses a kernel technique, in which the kernel is trained to capture mutual relations among different cell-lines, with respect to essentiality. We start with our baseline model, kernel ridge regression (KRR), a well-known, stably high-predictive performance model. We then attempt to improve the predictive performance of KRR by learning the kernel (or features) from gene essentiality data itself. More concretely we focus on the essentiality scores of genes, in given data, where the scores of different genes are heavily dependent on each other. We incorporate this dependency into our predictive model by using kernel canonical correlation analysis (KCCA) and kernel target alignment (KTA), both of which can be interpreted as estimating feature representations using the 'ideal' kernel defined by essentiality scores. After obtaining kernels through KCCA and KTA, we then predict the essentiality of an arbitrary gene by using the two KRR models. We finally take the average over the two prediction results (by the two models) to stabilize the results. An important point of our model is that the trained kernel is shared with all genes to predict the essentiality of each gene. This point reduces estimation variance, which can be a severe problem in high dimensional and small-sample data (which is applied to the given data this time), rather than estimating different kernels for each gene. Overall, these modifications make our predictive model a high-performance approach, particularly in subchallenge 1 of the Broad-DREAM Gene Essentiality Prediction Challenge. An additional, big advantage of our approach is computational efficiency, because all techniques (KCCA, KTA, and KRR) used in our approach are kernel methods, in which we do not have to deal with high dimensional data directly after we once calculate the kernels.
...............................................................................................................................
Tuesday, November 11
10:20 am – 10:40 am
DR T13
DREAM Best Performer Talk – GEP Challenge
A strategy to select most informative biomarkers for cancer cell lines
Fan Zhu1, Yuanfang Guan1
1University of Michigan
Cancer cells represent strong heterogeneity and thus the response to treatment varies dramatically between individuals. Currently, a rough estimation of 80% of the patients do not respond to cancer therapy. Personalized treatment of tumors thus requires accurate identification of drug targets for the specific samples collected from biopsy. Ideally, a test panel with a limited number of biomarkers can be designed for each type of cancer to identify effective drug targets for a patient. The Broad Institute Gene Essentiality Subchallenge 2 studies whether such biomarkers can be found for each type of cancer. We have developed a method to rigorously select such stable biomarkers based on both their informativeness in the cell line under investigation and the global informativeness over all cell lines. This was the best-performing method in this subchallenge.
...............................................................................................................................
Tuesday, November 11
10:40 am – 11:00 am
DR T14
DREAM Best Performer Talk – GEP Challenge
Predicting gene essentiality using linear-time greedy feature selection
Peddinti Gopalacharyulu*1, Alok Jaiswal*1, Kerstin Bunte2, Suleiman Khan1,2, Jing Tang1, Antti Airola4, Krister Wennerberg1, Tapio Pahikkala4, Samuel Kaski2,3, Tero Aittokallio1
*Equal contributions
1Institute for Molecular Medicine Finland FIMM, University of Helsinki, 2Helsinki Institute for Information Technology HIIT, Aalto University, 3Helsinki Institute for Information Technology HIIT, University of Helsinki, 4University of Turku
Genome-wide prediction of the gene essentiality using molecular characteristics of various cancer cells has the potential to open up new avenues for selective cancer therapies as well as for providing insights into the systems-level genetic interaction networks of cancer cells. Subchallenges 2 and 3 of the Broad-DREAM 9 Gene Essentiality Prediction Challenge deal with the problem of finding a limited number of molecular features that are most predictive of the gene essentiality. To solve this problem, we used a greedy forward feature selection algorithm for regularized least squares (RLS), called GreedyRLS. The GreedyRLS algorithm works like a wrapper type of feature selection method, which starts with an empty feature set, and in each iteration adds the feature whose addition provides the minimum RLS error in the leave-one-out cross-validation (LOO-CV). The GreedyRLS algorithm, however, performs the feature selection computationally more efficiently than previously known feature selection algorithms for RLS. The time complexity of the standard approach using LOO-CV for forward selection of k features from a total number of n features in a data set with m training samples is In contrast, the time complexity of the GreedyRLS is In subchallenge 3, we utilized the GreedyRLS approach for multi-task learning, and it performed the best among all the competing methods in this subchallenge. We addressed subchallenge 1 using additional information based on pathways from PARADIGM and gene sets from MSigDB. In this sub-challenge, we used the Bayesian multitask multiple kernel learning (BEMKL) method, which is a nonlinear method based on kernelized regression and Bayesian inference. Use of additional information of similarities of genes based on gene ontology seemed to be helpful in predicting gene essentiality, in line with the lessons learned from the previous NHI-DREAM Drug Sensitivity Prediction Challenge, but did not lead to the top performance in this subchallenge.


