|
With only one day left, ISMB/ECCB 2021 gave us a day starting off with Exhibitor Demos from EMBL-EBI, PerMedCoE, and GOBLET. The day continued into 9 different COSI tracks, including one for General Computational Biology, and a Special Session Final Project Presentations: Collaborative Tools for Protein Analysis Hackathon 2021.
Highlights
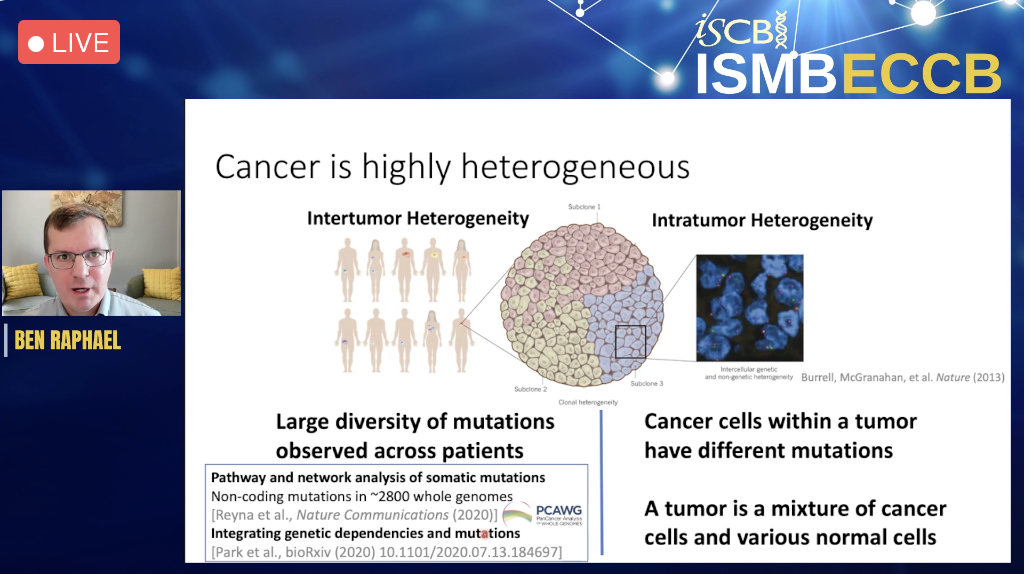
ISCB Innovator Award Keynote, Ben Raphael gave an engaging talk on Quantifying Tumor Heterogeneity across Time and Space. Tumors are heterogeneous mixtures of normal and cancerous cells with distinct genetic and transcriptional profiles. In this talk, he presented present several computational approaches to quantify tumor heterogeneity and to study tumor evolution using single-cell and spatial sequencing technologies. For single-cell DNA sequencing data, he described algorithms to reconstruct tumor evolution from multiple types of somatic mutations and how to use these approaches to analyze changes in tumor genomes over time. For spatial transcriptomics data, he introduced algorithms to detect genomic aberrations and to align and integrate data from multiple adjacent tissue sections leveraging both spatial and transcriptional similarity. The applications of these methods to quantify spatial heterogeneity in several cancer types were illustrated.
|
COSI Recaps
BOSC
BOSC 2021 (https://www.open-bio.org/events/bosc-2021/), the 22nd annual Bioinformatics Open Source Conference, kicked off this morning with opening remarks by BOSC Chair Nomi Harris, speaking from her home in California at 4:00am local time. The opening session also included an overview of the Open Bioinformatics Foundation by OBF President Peter Cock.
The first BOSC 2021 keynote talk, “Significant heterogeneities: Ecology’s emergence as open and synthetic science”, was delivered live by Christie Bahlai. Ecology has not been a big topic at past BOSCs, but Christie’s approach to open and inclusive science strongly resonated with our community. One attendee commented, “Christie Bahlai's keynote was inspiring and a breath of fresh air. +1 to calling out open science purists and the call for collective responsibility.”
Christie’s keynote was followed by a session on Standards and Practices for Open Science. One speaker in that session, Dhrithi Deshpande, spoke about the disappointingly low percentage of scientific papers that provide a link to the code (only about 12% -- though up from only 1% in 2016!), and noted that articles that share code tend to get cited more.
The next session, Tools for Open Science, started off with a well-received talk by Thorin Tabor about GenePattern, a reproducible research platform built on top of Jupyter Notebook.
Today’s final BOSC session was a joint session with Function COSI, chaired by Iddo Friedberg, featuring a keynote talk by Lara Mangravite on “Open approaches to advance data-intensive biomedicine.” 210 participants attended this joint session. Lara noted that clinical applications need access to high-quality data, but broad accessibility of human clinical data is difficult due to privacy issues, and access to analysis capabilities is distributed inequitably and tends to leave out those in the global south. Lara discussed some possible mitigations for those challenges.
Birds of a Feather, which bring together participants to chat informally about a shared interest, are always a popular feature of ISMB. In the BoF “Next steps for computational reproducibility toward fully executable papers”, there were discussions around ‘What is reproducibility?” and how do we incentivize new PIs to make reproducible work? Some cloud frameworks like Terra allow for reproducibility and lower the barrier for those who don’t have computational infrastructure in house. On the other hand, what about cloud costs? And is the reason why more papers aren’t reproducible the lack of skilled software engineers on the team?
The day ended with a hangout in the BOSC roundtable (which quickly ran into the 16-person maximum) that evolved into a discussion of the OBF and new ways for it to serve the open source bioinformatics community.
Don’t miss the second and final day of BOSC 2021 tomorrow, which begins with the last of our three keynotes, Thomas Hervé Mboa Nkoudou speaking about “Contribution of the maker movement to biotechnology in Africa: An open science perspective” (in French with English subtitles).
CAMDA
The 2nd day of virtual CAMDA 2021 started with an exceptional talk by Weida Tong, director of the Division of Bioinformatics and Biostatistics at the NCTR of the US FDA. Dr Tong discussed five common myths about AI and their implications for regulatory science, and provided a broad overview of how the FDA uses AI/ML techniques in support of the prediction of Drug Induced Liver Injury (DILI). This was followed by further presentations employing a variety of AI language models to identifying scientific papers relevant to drug induced liver injury, as benchmarked by a CAMDA challenge organized jointly with the US FDA with support of IARAI Vienna. The seond half of the day was devoted to the Metagenomic Phage Forensics of Anti-Microbial Resistance challenge based on recently published MetaSUB Consortium data, which was explored by the CAMDA community this year for the first time, yielding novel complex relationships between phages and AMR that indicated a new for further investigations. The rich world of modern meta genomic data was then brilliantly characterised in our keynote by Nikos Kyrpides from the DOE Joint Genome Institute, ranging from the Earth microbiome to the Global virome.
Join us on Friday for an introduction to Disease Maps for modelling COVID-19 by Maria Peña Chilet of the FPS in Seville, followed by contributed talks of the matching data analysis challenge. In the next session we host ISMB/ECCB proceedings presentations on using AI/ML in cancer studies as well as AI-driven Cloud Laboratories. We will wrap up the day with the CAMDA Cafe where the CAMDA community will discuss the Grand challenges of our times to tackle in the coming years, and finishing with the awards ceremony for the traditional CAMDA Trophy for the best presentation. Join us! See www.camda.info for details.
iRNA
The second day of the iRNA COSI 2021 was dominated by epitranscriptomics. The keynote was given by Kathi Zarnack from the Buchmann Institute for Molecular Life Sciences in Germany who discussed the detection/prediction of m6A RNA modifications using transcriptomics data and machine learning, identifying sites within consensus DRACH motifs but also others, providing a rich resource for further studies. Two contributed talks on splicing kinetics and modeling of multivalent binding by RNA-binding proteins were followed by five talks on RNA modification detection from sequencing, raising important common issues and challenges which were addressed in our live panel discussion, to which participated Nicole Martinez, Shengdong Ke and Schraga Schwartz. The panel highlighted the fact that while a decade ago, the challenge in epitranscriptomics was to determine where the modifications are, with rapid and considerable increases in technology capacity brought on by innovations in the field, we are now switching gears and focusing on questions like the function of the modifications, how to address the interconnectedness of the process and the effects throughout the life cycle of the targeted RNA, and the use of systematic perturbations and their analysis using machine learning to understand what the modifications are really doing in the cell. Our panel was followed by our second poster session and our traditional social time with quiz during which we took time to catch up and learned much trivia about the Olympics including surprising facts.
VarI
During the first day of the VarI meeting we had two keynote talks. Alessandra Carbone (Sorbonne University) presented her recent work on the analysis of evolutionary information for detecting protein sites in viral proteomes important for protein-protein interaction and drug resistance. Later, Ben Langmead (John Hopkins University) presented methods for reducing reference bias in the detection of human variations. The proposed approaches are based on new algorithms that allow to align sequencing reads to the reference genome of different populations. The VarI meeting also hosted a proceeding talk from Chirag Jain who presented a new approach for minimizing variation graph size that allows to reduce the computational complexity in the methods for reducing genome bias. In the first days the program included 6 selected presentations focusing on variant prioritization, sex dependent genotype association studies and pleiotropy. Finally, we had a poster session with 18 works presented.
Tomorrow we will have a keynote talk from Ben Lehner (Centre for Genomic Regulation) and a roundtable discussion with Yana Bromberg, (Rutger University), Douglas Fowler (University of Washington), Daniel Gilchrist (NHGRI) and Predrag Radivojac (Northeastern University). Alex Kaplun will present the research activity of Variantyx which has been our main sponsor in the last few years.
|
|
Cheat-Sheet Section/Platform Tips:
From time to time we may experience technical issues. When in doubt, give the system a quick refresh!
Updating the Time zone
The ISMB/ECCB 2021 virtual platform is enabled with time zone localization to enhance usability. In order to set the platform to your local time zone, click on "Full schedule" and then click on the dark grey "My Time Zone" box to ensure it is activated. Once clicked, the box will darken in color and your schedule will automatically be viewed in your local time zone.
Struggling with UTC conversion - UK +1 CEST +2 EDT -4, CDT -5, MDT -6, PDT -7, AEST +10, Tokyo +9, Beijing +8
|












