Art and Science Exhibition
ISMB/ECCB 2015 brings together scientists from a wide range of disciplines, including biology, medicine, computer science, mathematics and statistics. In these fields people are constantly dealing with information in visual form: from microscope images and photographs of gels to scatter plots, network graphs and phylogenetic trees, structural formulae and protein models to flow diagrams; visual aids for problem-solving are omnipresent. Some of the works of the first such exhibition at the
ISMB 2008 in Toronto combine outstanding beauty and aesthetics with deep insight that perfectly proves the validity of our approach or goes beyond the problem's solution. Others were surprising and inspiring through the transition from science to art, opening our eyes and minds to reflect on the work that we are undertaking.
The Art & Science Exhibition 2015 presents the artworks that have been generated as part of research projects. They are also soliciting images resulting from creative efforts that involve scientific concepts or employ scientific tools and methods.
Click image to download larger file
Art & Science Images |
A&S01
ALAA ABI Haidar
University of Pierre and Marie Curie, France
Discipline
Wormlike Light Reflections encapsulated in Soap Bubbles |  |
A&S02
ALAA ABI Haidar
University of Pierre and Marie Curie, France
Smilies
Soap + fats + water = smiling faces in Macro |  |
A&S03
Moshe Dessau
Bar-Ilan University, Israel
An artistic presentation of the glycoprotein C dimer
An artistic presentation of the glycoprotein C dimer from Rift valley fever virus. The two promotes (Red and Yellow) are in a molecular surface representation. |  |
A&S04
Moshe Dessau
Bar-Ilan University, Israel
An assembly model of Rift valley fever virus (RVFV)
The crystal structure of RVFV Gc, one of the virus' two envelope glycoproteins, fitted into a cryo-EM 3D reconstruction. (Dessau & Modis, PNAS 2013) |  |
A&S05
Boris Fichtman
Bar-Ilan University, Israel
SEM image of fruit fly (Drosophila melanogaster) hind foot.
SEM image of fruit fly (Drosophila melanogaster) hind foot. | 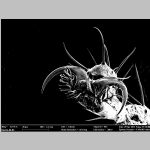 |
A&S06
Amnon Harel
Bar-Ilan University, Israel
Erythrocyte on placenta (human)
Erythrocyte on placenta (human) |  |
A&S07
Boris Fichtman
Bar-Ilan University, Israel
Glial cell from mice brain
Glial cell from mice brain | 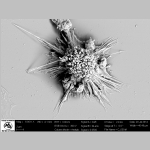 |
A&S08
Amnon Harel
Bar-Ilan University, Israel
cellular nuclei with nuclear pore complex
cellular nuclei with nuclear pore complex. | 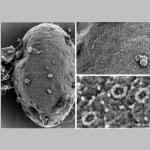 |
A&S09
Avraham Samson
Bar-Ilan University, Israel
Normal modes dynamics of multibody proteins: The acetylcholine receptor and its complex with snake toxins
Calculating protein dynamics is essential for a better understanding of many biological processes. To better understand the motion involved in channel opening of the acetylcholine receptor (AChR), I calculated the normal mode dynamics of the AChR (1). The normal modes calculations reveal a twist like gating motion responsible for channel opening. This motion is shown in the movie on the right. Throughout this twist motion, the ion channel diameter is shown to increase. Strikingly, the twist motion and the increase in channel diameter are not observed for the acetylcholine receptor in complex with two α-bungarotoxin (αBTX) molecules. The toxins seem to lock together neighboring receptor subunits thereby inhibiting channel opening. Interestingly, one αBTX molecule is sufficient to prevent the twist motion. These results shed light on the gating mechanism of the acetylcholine receptor, and present a complementary inhibition mechanism by snake venom derived α-neurotoxins. | 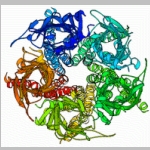 |
A&S10
Avraham Samson
Bar-Ilan University, Israel
Elastic network normal mode dynamics reveal the GPCR activation mechanism
G-protein-coupled receptors (GPCR) are a family of membrane-embedded metabotropic receptors which translate extracellular ligand binding into an intracellular response. Here, we calculate the motion of several GPCR family members such as the M2 and M3 muscarinic acetylcholine receptors, the A2A adenosine receptor, the β2 -adrenergic receptor, and the CXCR4 chemokine receptor using elastic network normal modes. The normal modes reveal a dilation and a contraction of the GPCR vestibule associated with ligand passage, and activation, respectively. Contraction of the vestibule on the extracellular side is correlated with cavity formation of the G-protein binding pocket on the intracellular side, which initiates intracellular signaling. Interestingly, the normal modes of rhodopsin do not correlate well with the motion of other GPCR family members. Electrostatic potential calculation of the GPCRs reveal a negatively charged field around the ligand binding site acting as a siphon to draw-in positively charged ligands on the membrane surface. Altogether, these results expose the GPCR activation mechanism and show how conformational changes on the cell surface side of the receptor are allosterically translated into structural changes on the inside | 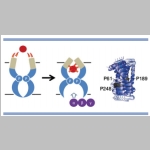 |
A&S12
Ryuta Nakajima
University of Minnesota Duluth, United States
Amburghese di cuore (no. 6)
Considering the salutations of contemporary visual culture, it is essential to investigate its’ effect and implication by asking such existential questions as Why do we make images, where do they come from and what is their primary function? In order to answer these rather difficult questions, this study focused on Sepia Pharaonis’s adaptive coloration as a empirical and biological model that codes and remaps visual information such as paintings, photographs, and video. More specifically, their adaptive coloration is triggered by both lateral and longitudinal visual stimuli replacing natural elements with various types of digital images. A LED flat-screen monitor controlled by a computer was placed directory under or next to a starphire-glass aquarium as a projection device. Through this study, we observed that basic camouflage tactics of uniform, mottled and disruptive pattern (Hanlon and Messenger 1996) are also applicable and viable methods in categorizing avant-garde paintings and photographs. In short, the body pattern produced by Sepia pharaonis can accurately reproduce culturally driven and highly conceptual images. The collected data from these experiments are used to create a series of photographs and videos installations. The genetically and evolutionally pure empirical may not only uncover certain key information needed to understand the origin of visual communication but also function as a catalyst that will redirect our culture away from the ever simulated hyper-reality. This, in tern, may create a truly valuable interdisciplinary platform to discuss the current trends in both art and science. |  |













