MAPS
Resort map (.pdf)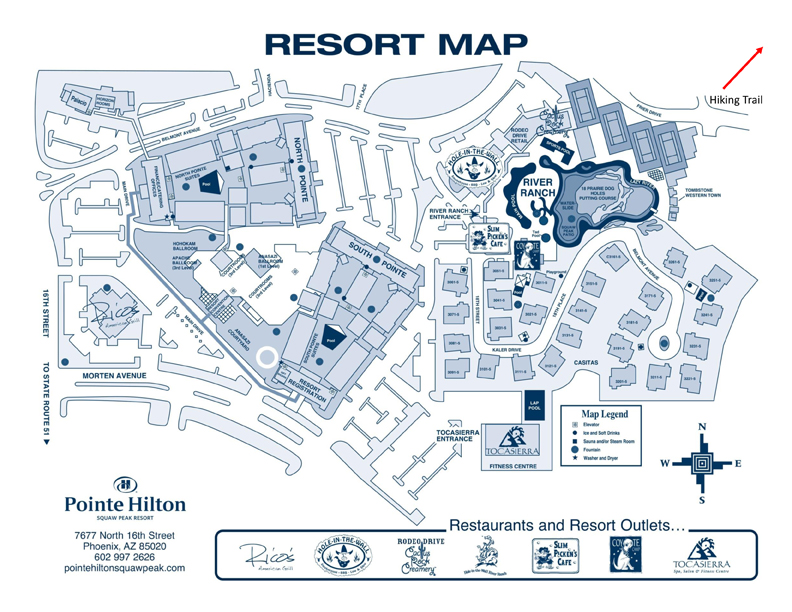
Resort Floor Plan (.pdf)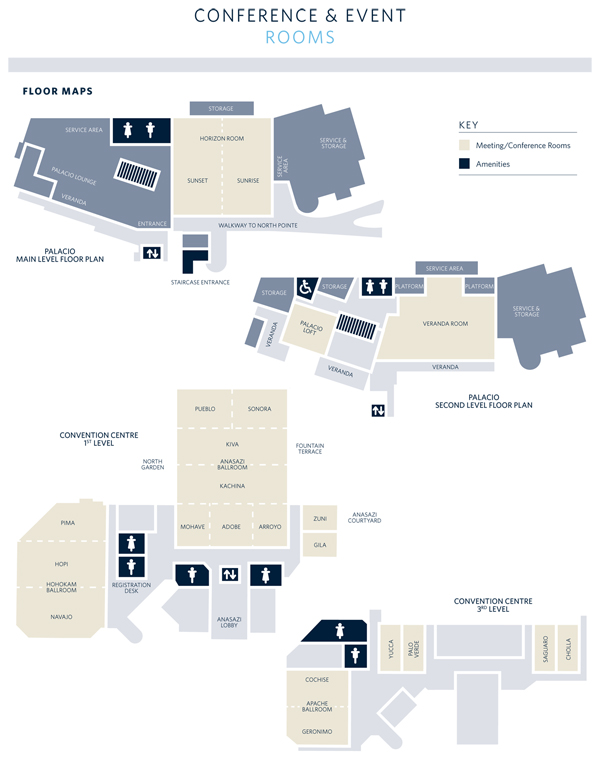
July 12 - 16, 2026
Washington, D.C., United States
ISCB Flagship Event
Dec 10-13, 2025
Hong Kong, China
ISCB Official Event
December 6 - 8, 2025
Northeastern University, USA
Jan 3 - 7, 2026
Big Island, Hawai'i
April 21-22, 2026
Cambridge, UK
ISCB Official Event
May 26 - 29, 2026
Thessaloniki, Greece
May 27-29, 2026
Toronto, Ontario, Canada
ISCB Official Event
August 31 - September 4, 2026
Geneva, Switzerland
Bioinformatics Community Events
ISCB’s Annual Flagship Meeting
Support the society while achieving your marketing goals
Become a ISCB collaborative conference, learn more here
Regional, topical, worldwide - your platform to present science
dedicated to facilitating development for students and young researchers
The ISCB Affiliates program is designed to forge links between ISCB and regional non-profit membership groups, centers, institutes and networks that involve researchers from various institutions and/or organizations within a defined geographic region involved in the advancement of bioinformatics. Such groups have regular meetings either in person or online, and an organizing body in the form of a board of directors or steering committee. If you are interested in affiliating your regional membership group, center, institute or network with ISCB, please review these guidelines (.pdf) and send your exploratory questions to Diane E. Kovats, ISCB Chief Executive Officer (This email address is being protected from spambots. You need JavaScript enabled to view it.). For information about the Affilliates Committee click here.
Topically-focused collaborative communities
Connect with ISCB worldwide
Environmental Sustainability Effort
ISCB is committed to creating a safe, inclusive, and equal environment for everyone
Resource library for education and training materials
Search jobs, find talent
Science at the click of the mouse, recorded talks
High-quality research devoted to computer-assisted analysis of biological data
Latest research and publications
Certifying Quality in Computational Biology Education
Latest updates from ISCB
Highlighting Society events, programs, and achievements
Celebrating scientific achievement and innovation
Honoring our distinguished researchers
Recognizing contributions and achievements
Center for science, collaboration, and training
Resort map (.pdf)
Resort Floor Plan (.pdf)
Link within this page: Download Mobile App
P# = Cytoscape related work
| Poster Number |
Authors | Title |
| P1 | Anastasia Baryshnikova | Functional Annotation of Large-Scale Biological Networks |
| P2 | Christopher Maher, Nicole White, Shuang Zhao, Jin Zhang, Emily Rozycki, Ha Dang, Sandra McFadden, Abdallah Eteleeb, Mohammed Alshalalfa, Ismael Vergara, Nicholas Erho, Jeffrey Arbeit, Jeffrey Karnes, Robert Den and Elai Davicioni | Integrative transcriptome analyses reveals PCAT14 promotes lethal prostate cancer |
| P3 | Yaron Orenstein, Lin Yang, Arttu Jolma, Yimeng Yin, Jussi Taipale, Ron Shamir and Remo Rohs | DNA Shape Readout Specificities of Different TF Families |
| P4 | Yaron Orenstein, Ryan Kim, Polly Fordyce and Bonnie Berger | Sequence libraries to cover all k-mers using degenerate characters |
| P5 | Mark Grimes, Nicolas Fernandez, Neil Clark, Avi Ma’ayan, Klarisa Rikova and Peter Hornbeck | Tandem Mass Tag Data Analysis Reveals Post-translational Modification Signaling Pathways in Lung Cancer Cell Lines |
| P6 | Daniela Börnigen, Francisco Ojeda, Stefan Blankenberg and Tanja Zeller | A genetic evaluation of multiple cardiovascular risk factors in the European population |
| P7 | Daniela Börnigen, Francisco Ojeda, Stefan Blankenberg and Tanja Zeller | Metabolomics analyses in cardiovascular diseases for biomarker discovery |
| P8 | Simon J. van Heeringen | Motif activity by ensemble learning for multi-experimental transcription factor motif analysis |
| P9 | Simarjeet Negi and Chittibabu Guda | Functional characterization of the healthy adult human brain and its application to study neurological disorders |
| P10 | Tamir Tuller | COP9 signalosome influences the epigenetic landscape of Arabidopsis thaliana |
| P11 | Alon Diament and Tamir Tuller | Inter-organismal analysis of Hi-C data demonstrates a relation between phenotypic divergence and gene re-organization |
| P12 | Florian Schmidt, Tzung-Chien Hsieh, Nina Gasparoni, Gilles Gasparoni, Wei Chen, Karl Nordström, Oliver Gorka, Jürgen Roland, Anupam Sinha, Philip Rosenstiel, Julia K. Polansky, Alf Hamann, Joern Walter and Marcel Schulz | A discriminatory method for the identification of transcriptional key regulators using epigenetic data |
| P13 | Krystelle Nganou Makamdop, Slim Fourati, Jianfei Hu, Amy Ransier, Farida Laboune, Rafick-Pierre Sekaly and Daniel Douek | Activation of IFNγ-signaling in myeloid dendritic cells is a new marker of risk of HIV-1 acquisition in the HVTN505 cohort |
| P14 | Kimberly Kanigel Winner and James Costello | A Spatiotemporal Model To Simulate Chemotherapy Regimens For Heterogeneous Bladder Cancer Metastases To The Lung |
| P15 | Heeju Noh, Ziyi Hua and Rudiyanto Gunawan | Inferring molecular targets and mechanism of action from time course expression data |
| P16 | Rahul Mohan, Michael Wainberg, Nasa Sinnott-Armstrong and Anshul Kundaje | kmer2vec: Unsupervised embedding of regulatory DNA sequences |
| P17 | Pascal Fieth, Gilles Monneret, Andrea Rau, Florence Jaffrezic, Alexander Hartmann and Gregory Nuel | Improving Causal Gaussian Bayesian Network Inference using Parallel Tempering |
| P18 | Morteza Chalabi Hajkarim | Computational Approaches to Decipher Composition and Regulation of Complexes by Large-scale Analysis of Mass Spectrometry (MS) Data |
| P19 | Zohar Zafrir and Tamir Tuller | Unsupervised detection of regulatory gene expression information in different genomic regions enables gene expression prediction |
| P20 | Hatice Osmanbeyoglu, Eneda Toska, Carmen Chan, Jose Baselga and Christina Leslie | Modeling the impact of somatic alterations on signaling pathways and transcriptional programs in human cancers |
| P21 | Brian Ross and James Costello | Modeling heterogeneous cell populations using Boolean networks |
| P22 | Xiangjia Min | Comprehensive cataloging and analysis of alternative splicing events in maize |
| P23 | Oana Ursu, Nathan Boley, Maryna Taranova, Rachel Wang and Anshul Kundaje | A graph diffusion framework for multi-scale reproducibility and differential analysis of 3D chromatin contact maps |
| P24 | Jayoung Kim | Interstitial Cystitis-Associated Urinary Metabolites |
| P25 | Michaela Willi and Lothar Hennighausen | Super-enhancers and CTCF in the regulation of mammary-specific genes |
| P26 | Avanti Shrikumar, Peyton Greenside, Anna Shcherbina, Johnny Israeli, Nasa Sinnott-Armstrong and Anshul Kundaje | Not Just a Black Box: Interpretable Deep Learning for Genomics |
| P27 | Mijin Kwon, Jinmyung Jung, Hasun Yu and Doheon Lee | Network-based inference of the effects of hormones on drug efficacy |
| P28 | Daniela Börnigen, Francisco Ojeda, Stefan Blankenberg and Tanja Zeller | Computational ten-steps workflow for large-scale metabolomics analysis in cardiovascular diseases |
| P29 | Alexendar Perez, Yuri Pritykin, Joana Vidigal, Sagar Chhangawala, Lee Zamparo, Christina Leslie and Andrea Ventura | GuideScan: a comprehensive and customizable guide RNA design tool |
| P30 | Istvan Ladunga, Avi Knecht, Adam Caprez, Timothy Bailey, Tao Liu, Pedro Madrigal and David Swanson | ChIPathlon: community-wide performance assessment of tools for mapping transcription factor binding sites and histone modifications |
| P31 | Hyunghoon Cho, Bonnie Berger and Jian Peng | Mashup: Compact Integration of Multi-Network Topology for Functional Analysis of Genes |
| P32 | Yongxiao Yang and Xinqi Gong | Protein-protein interface prediction: from several accurately predicted interface residue to the whole interface |
| P33 | Alberto Giaretta, Barbara Di Camillo, Luisa Barzon, Gianna Maria Toffolo and Timothy Charles Elston | Stochastic modeling of HPV early promoter gene regulation |
| P34 | Christopher Koch, Jay Konieczka, Toni Delorey, Ana Lyons, Amanda Socha, Kathleen Davis, Sara A Knaack, Dawn Thompson, Erin K O'Shea, Aviv Regev and Sushmita Roy | A phylogenetic framework to study the evolution of transcriptional regulatory networks |
| P35 | Alireza Fotuhi Siahpirani and Sushmita Roy | A prior-based integrative framework for functional transcriptional regulatory network inference |
| P36 | H. Tomas Rube, Chaitanya Rastogi and Harmen J. Bussemaker | Revisiting the relationship between DNA shape, base sequence, and transcription factor binding |
| P37 | Qi Song and Song Li | CoReg: Identification of co-regulators in genome scale transcription regulatory networks |
| P38 | Xuewei Wang, Zhifu Sun, Jean-Pierre Kocher and Burgrim Andrej | Pathway-based approach to predict drug response of cancer cells |
| P39 | Khader Ghneim, Dan Barouch, Jeffrey Lifson and Rafick-Pierre Sekaly | Rapid Inflammasome Activation following Mucosal SIV Infection of Rhesus Monkeys |
| P40 | Cynthia Kalita, Gregory Moyerbrailean, Christopher Brown, Francesca Luca and Roger Pique-Regi | A novel allele-specific data analysis method for high-throughput reporter assays |
| P41 | Huy Pham and Alioune Ngom | A new effective feature selection method and its application for classification on biological data |
| P42 | Lingfei Wang and Tom Michoel | Scalable causal gene network inference from genetics of gene expression data |
| P43 | Gil Speyer, Jeff Kiefer, Hai J. Tran, Harshil Dhruv, Michael Berens and Seungchan Kim | Identification of personalized therapeutic targets via single-cell transcriptomic analysis |
| P44 | Anasuya Pal, Jia Zeng, Laura Gonzalez-Malerva, Seron Eaton, Jin Park and Joshua Labaer | Integrated pathway analysis on phenotypic and molecular alterations induced by different missense mutant p53 proteins in mammary epithelial cells |
| P45 | Yungil Kim, Xin Li, Emily Tsang, Joe Davis, Farhan Damani and Colby Chiang | The impact of rare variation on gene expression across tissues |
| P46 | Casey Hanson, Junmei Cairns, Liewei Wang and Saurabh Sinha | A Probabilistic Graphical Model integrates genotype, gene expression and ChIP-seq data to identify transcription factors associated with drug response variation |
| P47 | Robert Vogel, Mehmet Eren Ahsen and Gustavo Stolovitzky | Unsupervised aggregation of rank predictions for binary classification in translational systems biology tasks |
| P48 | Ning Shen, Jingkang Zhao and Raluca Gordan | Comparing the specificity of siblings in transcription factor families |
| P49 | Andrea Repele, Shawn Krueger and Manu Manu | Experimental test of a novel de novo regulatory decoding methodology in the context of the Cebpa locus |
| P50 | Michael Saul, Wei Yang, Laura Sloofman, Abbas Burkhari, Charles Blatti, Chris Seward, Joe Troy, Hagai Shpigler, Sriram Chandrasekaran, Alison Bell, Gene Robinson, Lisa Stubbs, Dave Zhao and Saurabh Sinha | Cross-species network analyses reveal conserved genomic toolkits involved in response to social challenge |
| P51 | Emily Miraldi, Maria Pokrovskii, Jason A. Hall, Dayanne Martins de Castro, Nicholas Carriero, Aaron Watters, Ren Yi, Dan R. Littman and Richard Bonneau | Learning how diverse gut immune cell types contribute to autoimmune disease through reverse-engineered transcriptional regulatory networks |
| P52 | Mehmet Eren Ahsen, Amanda Craig, Carlos Villacorta-Martin, Xintong Chen, Ismail Labgaa, Ashley Stueck, Delia D’avola, Stephen Ward, Maria Isabel Fiel, Ganesh Gunasekaran, Josep Llovet, Swan Thung, Myron Schwartz, Bojan Losic, Augusto Villanueva and Gustavo Stolovitzky | Multi-regional integrative genomic analysis reveals intra-tumor heterogeneity in a subset of early-stage hepatocellular carcinoma |
| P53 | Ana Stanescu and Gaurav Pandey | Learning parsimonious ensembles for biomedical prediction problems and DREAM challenges |
| P54 | Chen Chen, Min Zhang and Dabao Zhang | A Parallel Algorithm to Construct the Entire Gene Regulatory Network of an Organism |
| P55 | Johnny Israeli, Nathan Boley and Anshul Kundaje | Integrative deep regulatory genomic neural networks for predicting in-vivo transcription factor binding sites |
| P56 | Chenyue Hu, Alex Bisberg and Amina Qutub | Visually Guided Clustering: an integration of semi-supervised clustering with interactive visualization |
| P57 | Chenyue Hu, Steven Kornblau and Amina Qutub | Meta-Proteomic Analysis for the Clinic: A Guide Towards Personalized Therapy in Leukemia |
| P58 | Allison Richards, Athma Pai, Daniel Kurtz, Gregory Moyerbrailean, Omar Davis, Chris Harvey, Adnan Alazizi, Yoram Sorokin, Nancy Hauff, Xiaoquan Wen, Roger Pique-Regi and Francesca Luca | Gene expression and splicing responses to environmental perturbations across 250 cellular conditions |
| P59 | Yan Kai, Alexandros Tzatsos and Weiqun Peng | Predicting CTCF-mediated chromosomal long-range interactions using genomic and epigenomic features |
| P60 | Victor Solovyev and Ramzan Umarov | Prediction of Prokaryotic and Eukaryotic Promoters Using Convolutional Deep Learning Neural Networks |
| P61 | Madhavi Ganapathiraju, Srilakshmi Chaparala and Sandeep Subramanian | DNA Palindrome variants and their association with diseases |
| P62 | Xi Gu and Chung-Jui Tsai | Gene co-expression analyses of parasitic plants reveal non-photosynthetic functions of phylloquinone in transmembrane parasitic signaling |
| P63 | Artur Jaroszewicz and Jason Ernst | Modeling of spatial organization of chromatin states facilitates discovery of various types of exon splicing patterns |
| P64 | Sebastian Winkler and Oliver Kohlbacher | Identifying deregulated subnetworks bridging different omics-layers via fractional and bi-level integer programming |
| P65 | Shun-Yuen Kwan, Jia-Hsin Huang, Zing Tsung-Yeh Tsai and Huai-Kuang Tsai | Expansion of transcription binding specificities in the promoters of humans |
| P66 | Ying Liu | Discovering Structures in Biological networks using pattern-based network decomposition |
| P67 | Tsu-Pei Chiu and Remo Rohs | Genome-wide prediction of minor groove electrostatics enables biophysical modeling of protein-DNA binding |
| P68 | Tunca Dogan, Ece Akhan, Marcus Baumann, Ahmet Süreyya Rifaioğlu, Diego Poggioli, Andrew Nightingale, Ian R. Baxendale, Maria Martin and Rengul Cetin-Atalay | Automated compound-protein interaction estimation and validation on PI3K/AKT/mTOR pathway |
| P69 | Anthony Findley, Gregory Moyerbrailean, Francesca Luca and Roger Pique-Regi | Transcription factor footprints predict the gene expression response to environmental stimuli |
| P70 | Jianwen Fang | Tight Integration of Multi-Omics Data Using Tensor Decomposition |
| P71 | Abiel Roche-Lima, Kelvin Carrasquillo-Carrion, Pablo Vivas and Dayanara Velazquez-Morales | Associating miRNA and Cancer in Hispanics Populations using Bioinformatics Analysis |
| P72 | Andrew Rouillard, Mark Hurle and Pankaj Agarwal | Selection of Features from Omic Datasets for Prioritizing Drug Targets |
| P73 | Tristan Bepler, Ryan Chung and Bonnie Berger | Protein secondary structure annotation using long-short term memory recurrent neural networks |
| P74 | George Steinhardt and David Waxman | PREDICTION OF TRANSCRIPTIONF FACTOR BINDING SITES USING ATAC-SEQ AND DNASE-SEQ. |
| P75 | Luis Santos, Robert Vogel, Marc Birtwistle, Jerry Chipuk, Pablo Meyer and Gustavo Stolovitzky | Mitochondria variability confers resilience to pro‐apoptotic stimulation and is associated with drug resistance |
| P76 | Gracia Bonilla and David Waxman | Chromatin state preferences of transcription factors in male and female mouse liver |
| P77 | Rosario Corona, Emily Adler, Janet Lee, Kate Lawrenson, Simon Gayther and Dennis Hazelett | Characterization of the Pax8 regulatory network in epithelial ovarian cancer |
| P78 | Padmapriya Swaminathan, Anne Fennell, Kalley Besler, Yuxin Yao, Somchai Rice, Jacek Koziel, Murli Dharmadhikari, Devin Maurer, Emily Del Bel, Zata Vickers, Katie Cook, Adrian Hegeman and Jim Luby | Characterization of Grape berry ripening - Genomics to Sensory |
| P79 | Simon Coetzee, Zack Ramjan, Huy Dinh, Benjamin Berman and Dennis Hazelett | StateHub-StatePaintR: a tool for creating and documenting reproducible chromatin state annotations. |
| P80 | Prathik Naidu and Caleb Lareau | A Machine Learning Approach to Identify Genome-Wide 3D DNA Interactions |
| P81 | Satyanarayan Rao, Harmen Bussemaker and Remo Rohs | High-throughput predictions of DNA shape features for methylated DNA reveal mechanisms of TF binding |
| P82 | David Tanenbaum Phd, Eldred Ribeiro Phd, Wenling Chang Phd, Peter Gutgarts, Ari Abrams-Kudan, William Kim, Lisa Tutterow, Lauren Quattrochi Phd and Erin Williams Jd | Catalyzing Biomedical Research through the NIH Commons Credit Cloud Computing Paradigm |
| P83 | Jared Sagendorf, Helen Berman and Remo Rohs | DNAproDB: A Web-Based Database and Visualization Tool for the Analysis of DNA-Protein Complexes |
| P84 | Murat Cokol and Bree B. Aldridge | Efficient and robust measurement of high order drug interactions |
| P85 | Christopher Michael Pietras, Diana W. Bianchi, and Donna K. Slonim | TEMPO: Temporal Modeling of Pathway Outliers |
 Download the RECOMB/ISCB RSG with Dream 2016 MOBILE APP!
Download the RECOMB/ISCB RSG with Dream 2016 MOBILE APP!
SEARCH: The App Store or Google Play for ISCB Conferences
SCAN:

Link within this page: Download Mobile App
D# = Cytoscape related work
| Poster Number |
Authors | Title |
| D1 | Istvan Ladunga, Avi Knecht, Adam Caprez, Timothy Bailey, Tao Liu, Pedro Madrigal and David Swanson | ChIPathlon: community-wide performance assessment of tools for mapping transcription factor binding sites and histone modifications |
| D2 | Ana Stanescu and Gaurav Pandey | Learning parsimonious ensembles for biomedical prediction problems and DREAM challenges |
| D3 | Robert Vogel, Mehmet Eren Ahsen and Gustavo Stolovitzky | Unsupervised aggregation of rank predictions for binary classification in translational systems biology tasks |
| D4 | David Tanenbaum Phd, Eldred Ribeiro Phd, Wenling Chang Phd, Peter Gutgarts, Ari Abrams-Kudan, William Kim, Lisa Tutterow, Lauren Quattrochi Phd and Erin Williams Jd | Catalyzing Biomedical Research through the NIH Commons Credit Cloud Computing Paradigm |
| D5 | Dennis Wang, Michael P. Menden, Michael Mason, Thomas Yu, Krishna C. Bulusu, Elias Chaibub Neto, In Sock Jang, Zara Ghazoui, John Vincent, Eric Tang, Giovanni Di Veroli, Gustavo Stolovitzky, Jonathan R. Dry, Justin Guinney, Julio Saez-Rodriguez | Community analysis to predict therapeutic synergy within the AstraZeneca-Sanger Drug Combination Challenge |
| D6 | Minji Jeon, Sunkyu Kim, Sungjoon Park, Heewon Lee, Hyeokyoon Chang, Minhwan Yu, Kwanghun Choi, and Jaewoo Kang | Drug Synergy Prediction using High Performance Computing and Support Vector Regression |
| D7 | Tin Nguyen, Bence Szalai, Gábort Turu, Miklós Cserző, László Hunyady, Sorin Draghici, Russ Wolfinger | Boosted Tree Based Prediction of Drug Synergisms in the AstraZeneca-Sanger Drug Combination Prediction DREAM Challenge |
| D8 | Weijia Zhang, Thuc Le, Taosheng Xu, Junpeng Zhang, Lin Liu, Jiuyong Li | Disease module identification with balanced multi-layer regularized Markov cluster algorithm |
| D9 | Chi W Pak | Bootstrap feature selection of early differentially expressed genes in human respiratory viral challenge studies: viral shedding and patient symptoms |
| D10 | Maria Suprun, Joel Correa-da-Rossa, Mayte Suarez-Farinas and Lewis E Tomalin | Estimating a generalizable ‘final model’, suitable for clinical classification applications, using Monte Carlo resamples of patient data |
| D11 | Artem Lysenko, Piotr J. Kamola, Keith A. Boroevich and Tatsuhiko Tsunoda | Strategies for discovering disease-associated modules in integrated biological networks |
| D12 | Emilie Ramsahai and Melford John | M-Reach pathways from networks combined on V-Structures |
| D13 | Jake Crawford, Junyuan Lin, Xiaozhe Hu, Benjamin Hescott, Donna Slonim and Lenore J. Cowen | A double spectral approach to the DREAM module identification challenge |
| D14 | Sergio Gómez, Manlio De Domenico and Alex Arenas | Module Identification by Adjusting Resolution in Community Detection Algorithms |
| D15 | Shana White, Lixia Zhang and Mario Medvedovic | Application of diffusion kernels of network graphs and subsequent clustering approaches for identification of disease modules |
| D16 | Hongjiu Zhang and Yuanfang Guan | An ultra-fast tumour heterogeneity inference method using density-based infinite mixture model |
| D17 | Liye He, Eemeli Leppäaho, Motoki Shiga, Bhagwan Yadav, Jussi Gillberg, Suleiman Khan, Zia ur Rehman, David Tamborero, Pekka Marttinen, Mehmet Gönen, Hiroshi Mamitsuka, Krister Wennerberg, Samuel Kaski, Tero Aittokallio, and Jing Tang | Target-based ensemble drug combination predictions for the DREAM10 challenge |
| D18 | Eduardo G. Gusmao, Zhijian Li, Ivan G. Costa | Prediction of Active Transcription Factor Binding Sites Using Computational Footprinting Data |
| D19 | Andrey S. Lando, Ilya E. Vorontsov, Valentina Boeva, Grigory V. Sapunov, Irina A. Eliseeva, Vsevolod J. Makeev, and Ivan V. Kulakovskiy | Preselection of training cell types improves prediction of transcription factor binding sites |
| D20 | Zafer Aydın | Discovering Biomarkers for Early Prognosis of Respiratory Virus Infection |
| D21 | Sandeep Kumar Dhanda, Kumardeep Chaudhary, Ritesh Kumar, Deepak Singla | ViResPred: A Solution for Respiratory Viral Dream Challenge |
| D22 | Michał Łopuszyński and Miron Bartosz Kursa | The application of the feature hashing and the mutual information based feature filter in predicting susceptibility to respiratory viruses |
 Download the RECOMB/ISCB RSG with Dream 2016 MOBILE APP!
Download the RECOMB/ISCB RSG with Dream 2016 MOBILE APP!
SEARCH: The App Store or Google Play for ISCB Conferences
SCAN:

As selected at RECOMB/ISCB Regulatory and Systems Genomics 2016
(Papers are listed alphabetically by title)
A widespread role of the motif environment in transcription factor binding across diverse protein families, Dror I, Golan T, Levy C, Rohs R, Mandel-Gutfreund Y. Genome Research 2015 Sep;25(9):1268-80
Affinity regression predicts the recognition code of nucleic acid-binding proteins, Pelossof R, Singh I, Yang JL, Weirauch MT, Hughes TR, Leslie CS. Nature Biotechnology 2015 Dec;33(12):1242-1249
Analysis of computational footprinting methods for DNase sequencing experiments, Gusmao E, Allhoff M, Zenke M, Costa I. Nature Methods 2016 Apr;13(4):303-9
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks, Kelley DR, Snoek J, Rinn J. Genome Research 2016 Jul;26(7):990-9
Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes, Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A, McKenna D, Stamenova EK, Lander ES, Aiden EL. Proc Natl Acad Sci U S A 2015 Nov 24;112(47):E6456-65
Evaluating the impact of single nucleotide variants on transcription factor binding, Shi W, Fornes O, Mathelier A, Wasserman WW. Nucleic Acids Research 2016 Aug 4. pii: gkw691
Inferring causal molecular networks: empirical assessment through a community-based effort, Hill SM, Heiser LM, Cokelaer T, Unger M, Nesser NK, Carlin DE, Zhang Y, Sokolov A, Paull EO, Wong CK, Graim K, Bivol A, Wang H, Zhu F, Afsari B, Danilova LV, Favorov AV, Lee WS, Taylor D, Hu CW, Long BL, Noren DP, Bisberg AJ; HPN-DREAM Consortium, Mills GB, Gray JW, Kellen M, Norman T, Friend S, Qutub AA, Fertig EJ, Guan Y, Song M, Stuart JM, Spellman PT, Koeppl H, Stolovitzky G, Saez-Rodriguez J, Mukherjee S. Nature Methods 2016 Apr;13(4):310-8
Integrating Transcriptomic and Proteomic Data Using Predictive Regulatory Network Models of Host Response to Pathogens, Chasman D, Walters KB, Lopes TJ, Eisfeld AJ, Kawaoka Y, Roy S. PLoS Computational Biology 2016 Jul 12;12(7):e1005013
Models of human core transcriptional regulatory circuitries, Saint-Andre V, Federation AJ, Lin CY, Abraham BJ, Reddy J, Lee TI, Bradner JE, Young RA. Genome Research 2016 Mar;26(3):385-96
Pathways on demand: automated reconstruction of human signaling networks, Ritz A, Poirel CL, Tegge AN, Sharp N, Simmons K, Powell A, Kale SD, Murali TM. Npj Systems Biology And Applications, vol. 2 p. 16002.
CONGRATULATIONS TO ALL AUTHORS!
Please check back for updates.
Please check back for updates.
Cytoscape Workshop
Sunday, November 6, 2016
1:00 pm - 5:00pm
- Registration Fees for Cytoscape Workshop (Click here)
The network perspective on biology aims to bring meaningful context to high-throughput data for exploratory analysis, interpretation and hypothesis generation. As a free and open source tool, Cytoscape has become the most popular network visualization and analysis tools in the biological sciences. It is now cited in over 600 publications per year and downloaded 14,000 times per month. This workshop will provide a general introduction to network biology studies and Cytoscape concepts, including a hands-on session for universal data import and demonstration of a few of the over 250 freely available apps contributed by the Cytoscape developer community.
By the end of this workshop, you should be able to:
-Import any tabular data into Cytoscape
-Integrate your data with public sources of networks, pathways and other datasets
-Master network layout and data visualization
Interested in participating in this workshop? Don't forget to register.
ISCB Members enjoy discounts on conference registration (up to $150), journal subscriptions, book (25% off), and job center postings (free).
Connecting, Collaborating, Training, the Lifeblood of Science. ISCB, the professional society for computational biology!
Giving never felt so good! Considering donating today.