Q: What is ISCB?
A: The International Society for Computational Biology (ISCB) is the parent organization of the annual Great Lakes Bioinformatics (GLBIO) Conference. The ISCB is dedicated to advancing the scientific understanding of living systems through computation. The ISCB communicates the significance of our science to the larger scientific community, governments, and the public at large. The ISCB serves a global membership by impacting government and scientific policies, providing high quality publications and meetings, and through distribution of valuable information about training, education, employment and relevant news from related fields. ISCB membership offers many benefits including reduced conference registration fees to several high impact events and reduced subscription prices for a selection of journals of Computational Biology and Bioinformatics. Members are from more than 50 countries and include over 800 students and nearly 500 post doctoral researchers.
- top -
Q: Who do I contact for information on the conference?
A: You can get information on the conference from:
Bel Hanson
ISCB Director of Operations & Programs
Phone: +1-571-293-2113
Email: This email address is being protected from spambots. You need JavaScript enabled to view it.
Justine Morris
ISCB Coordinator of Operations & Programs
Phone: +1-571-293-2113
Email: This email address is being protected from spambots. You need JavaScript enabled to view it.
- top -
Q: Where will the conference be held?
A: The conference will be held at the University of Pittsburgh:
University of Pittsburgh
4200 Fifth Avenue
Pittsburgh, Pennsylvania (PA)
15260
www.pitt.edu
- top -
Q: Is there a map of the venue?
A: Yes, you can download a PDF here.
- top -
Q: What Hotel is being used for the conference?
A: The GLBIO organizers have multiple conference hotel rooms and discounted rates available for rooms close to the conference venue - for details visit this link.
- top -
Q: Are Children allowed at the conference?
A: ISCB conferences are professional events. Children of registered ISCB conference attendees are welcome to attend the conference with their parent or guardian, as long as younger children are under the supervision of a parent or guardian at all times. Parents or guardians may bring children to educational events provided the child does not disrupt the event.
- top -
Q: Will child care be available at?
A: Yes, child care arrangements will be available through a professional child care service - more details will be available soon on how to register for this service.
- top -
Q: How do I get from the airport to my hotel? How can I travel around the city?
A: Pittsburgh Regional Transit (PRT) is the public transit agency for the greater Pittsburgh area, providing a variety of transportation options including bus, light rail, incline and paratransit service. You can find more information here.
Uber and Lyft are also popular services in the area and can provide you with an affordable ride. Cab Services and Parking Tips can be found in this additional link here. An accessibility guide if needed can be found here.
The two hotels in our block both have parking that can be added to your reservation (this can occur when you arrive and does not need to be pre-arranged). If you are not staying at one of those two properties, you can view other parking options here.
The closest airport to the University of Pittsburgh is Pittsburgh International Airport, which is approximately 20 miles (32 kilometers) away.
- top -
Q: What attractions and sightseeing opportunities are available?
A: Pittsburgh is located in western Pennsylvania at the junction of 3 rivers. It is a unique, vibrant city with a small-town feel but a wide variety of wonderful sights and attractions to enjoy. These include Arts & Culture, Sports, Annual Events & Festivals, Celebrations of Diversity, and even Free Events!
A full visitor’s guide can be found here.
And here are some fun facts about Pittsburgh to read at your leisure!
- top -
Q: What restaurants are close to the venue or my hotel?
A: You can find a list here. When searching, you can click ‘Near Address’ under the ‘Sort By’ heading and find what is closest to a specific location, or click ‘Near Me’ and it will use your GPS location. You can also search by keyword or cuisine.
- top -
Q: Are there any special discounts available for attendees?
A: VisitPittsburgh has created the “Show Your Badge” program for conference visitors to enjoy offers and discounts from partnered businesses and attractions. You can find more information on that program here.
- top -
Q: What is the local Time Zone?
A: Eastern Standard Time (EST) applies in Pennsylvania, USA.
- top -
Q: What is the climate?
A: The month of May in Pittsburgh has an average low temperature of 52°F (11°C) and an average high temperature of 72°F (22°C). Attendees should consider bringing a light jacket or raincoat in case of spring rain showers.
- top -
Q: Where can I find medical or emergency information?
A: More than 500 emergency phones are available throughout the Pittsburgh campus. Many phones are located outdoors and identified by a blue light. Once you press the phone's red emergency button, it will automatically register your location and connect you directly to University police.
You can also contact University Police via 412-624-2121 (emergencies) or 412-624-4040 (non-emergencies). For all medical emergencies, fire emergencies, or situations requiring the Pittsburgh Bureau of Police, please call 911.
All phone numbers are also listed here.
Q: How can I pay to register?
A: Delegates can pay on line during registration using a Visa, MasterCard or American Express credit card. Delegates wishing to pay using a wire transfer would create an invoice at time of registration and then contract the conference registrar for instructions.
- top -
Q: Where do I register when I arrive?
A: Conference Registration is available from Monday, May 13th to Thursday, May 16th at the University of Pittsburgh.
REGISTRATION DESK HOURS – May 13 – 16
University of Pittsburgh - Watch for signage
|
Monday, May 13
|
7:30 AM – 5:30 PM
|
|
Tuesday, May 14
|
8:00 AM – 5:00 PM
|
|
Wednesday, May 15
|
8:00 AM – 5:00 PM
|
|
Thursday, May 16
|
8:00 AM – 3:00 PM
|
- top -
Q: What should I do if I lose something at the conference venue?
A: A conference lost and found will be located at the conference information booth located at the University of Pittsburgh.
- top -
Q: What is the policy of photography or image capture?
A: ISCB encourages open sharing of science. As an author, you have the right to restrict the use of photography or any other form of capture of your research. If you do not wish to allow others to share your research, simply disclose this at the start of your oral presentation or add a note to your poster.
- top -
Q: What are the hashtags for the conference?
A: The hashtag is #GLBIO or #GLBIO2024. @ISCB is the ISCB Twitter/X handle.
We strongly encourage the use of Twitter/X at the conference to disseminate information. ISCB kindly requests that you observe the Conference Code of Conduct when tweeting. If you have a specific complaint, suggestion, or comment related to conference logistics, audio, heating and cooling, bathroom cleanliness, etc, we kindly request that you communicate your wish to a volunteer wearing an ISCB shirt, go to the conference registration desk, or an ISCB team member. Due to limited resources and the demands of managing the conference ISCB is unable to monitor Twitter/X fully and may miss an improvement request.
- top -
Click Schedule to download pdf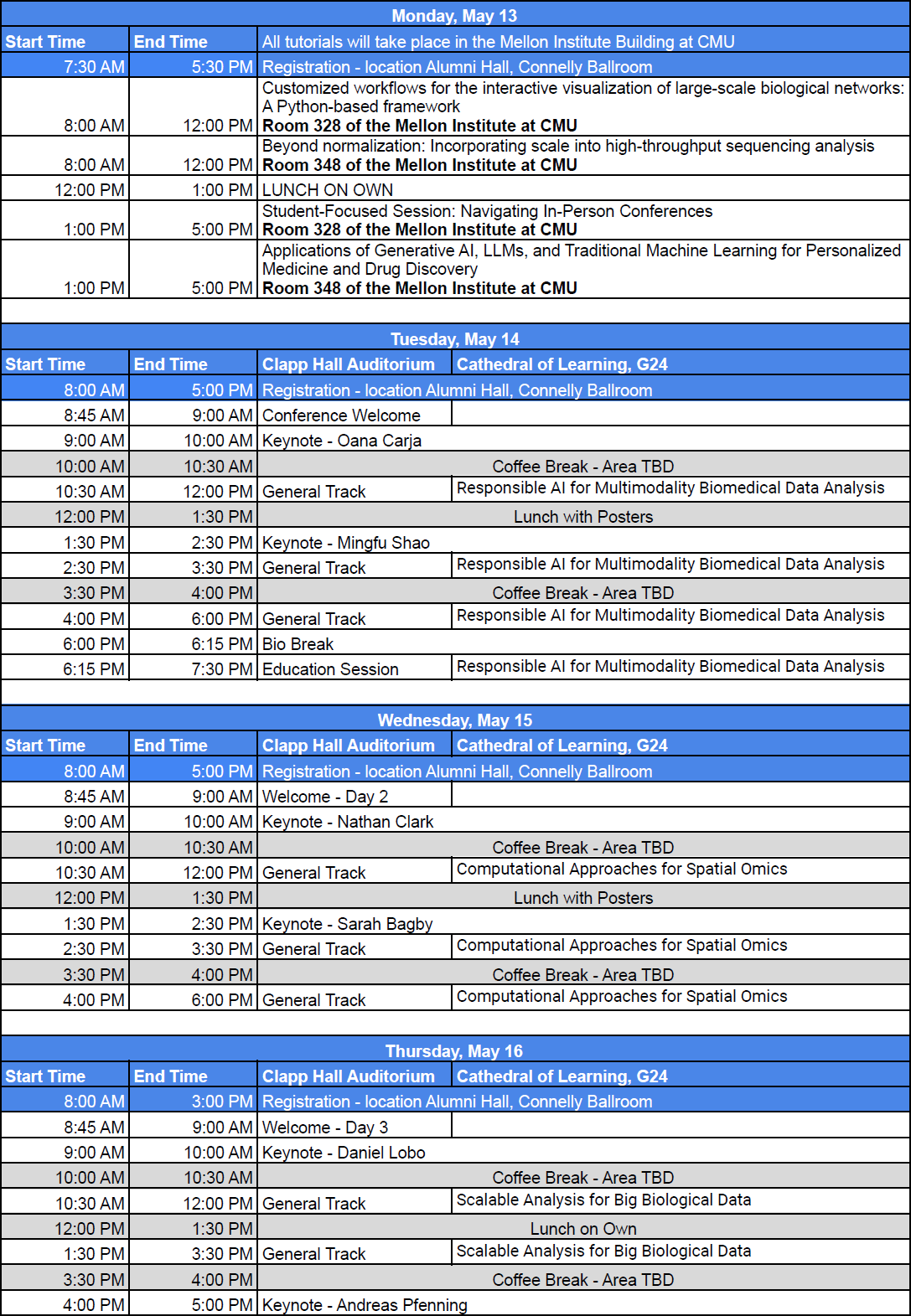
- top -
Links within this page: Sponsor | Funding Information | Travel Fellowship Application Overview | Maximum Awards and Amounts | Eligibility Requirements | Application Process | Eligible Expenses | Notification | Contacts
Sponsors



- top -
Funding Information
ISCB, through funding provided by the National Science Foundation, the International Society for Computational Biology, and the Great Lakes Bioinformatics Consortium are pleased to offer travel fellowships for students to attend GLBIO 2023 in Montreal, Quebec. Students from the United States, Canada, and other countries are encouraged to apply but funding sources for Travel Fellowships are very limited and we regret that we are not able to fund all applicants. The conference organizers are committed to providing support to as many eligible applicants as possible. Travel Fellowship consideration is based on criteria listed below.
- top -
Travel Fellowship Application Overview
- Applicant must be a current student (high school, undergraduate or graduate) to submit an application.
- All applicants must attend all three (3) conference days and secure additional funding from other sources in order to be able to cover the full costs of attending the conference.
- The deadline to submit a fellowship application is March 27, 2023, by 11:00 pm Eastern Daylight Time. No exceptions will be made.
|
Travel Fellowship Key Dates
|
|
March 27, 2023
|
Travel Fellowship Application Deadline (11pm Eastern Daylight Time)
|
|
April 7, 2023
|
Travel Fellowship Acceptance Notification
|
|
April 12, 2023
|
Early Registration Deadline (11:00 pm Eastern Daylight Time)
|
- top -
Maximum Award Amounts
The maximum fellowship award is $500 USD. Please note that funded applicants will only be able to cover approximately 50% of the expense of travel and registration fees with these fellowship amounts. Thus all applicants must seek and secure additional funding sources (e.g., from your home institution/university, or grant funding) and a letter regarding this additional funding is required in PDF format as part of your application.
- top -
Eligibility Requirements
- You are a student High School or registered in an undergraduate or graduate degree program. Postdocs and Professionals DO NOT qualify. Evidence of your student status must be presented to the onsite registration desk upon check-in at the conference.
- You will attend all three days of the conference, with arrival no later than the morning of May 16 and departure no earlier than the afternoon of May 18, and will sign in at the registration desk on each of these mornings to verify your attendance
- Applicant must be prepared to register for GLBIO 2023 by the early registration deadline of April 12, 2023. If attendance at the conference is dependent on receipt of travel fellowship funds, please do not register until after the notification of travel fellowship funding. Any funded applicant failing to register for the conference by the early registration deadline of April 12, 2023 will automatically forfeit the funds so that another applicant can be awarded from among the original pool of applicants.
- You have other funding resources to cover costs not met by an ISCB travel fellowship award. Applicant must be able to pay all expenses of attending the conference up front, including conference registration fee (as noted above), travel, accommodations and meals. Travel Fellowship funding will be provided as a reimbursement as a check payable in USD and mailed to the participant approximately 6-8 weeks after the conference.
- top -
Application Process
Please complete the application form:
No application will be accepted after the deadline of March 27, 2023. Special consideration will be given to first-time GLBIO attendees who are not necessarily presenting work at the conference, women, undergraduate or high school students, underrepresented minorities, or students with disabilities.
- top -
Eligible Expenses
Eligible expenses toward travel fellowship funds include conference registration, transportation (air on a U.S. Flag air carrier or land transportation from home region to conference city), and a maximum of 250 USD of hotel room and occupancy tax charges. In order to receive the full-awarded amount, receipts for registration, transportation and hotel accommodations are required that equal or exceed the awarded amount. Conference registration fee is reimbursable at the early registration pricing only; registration fees to attend pre-conference events are not reimbursable expenses.
- top -
Notification
Applicants will be notified no later than April 7, 2023 of the funding status. In some cases applicants may be notified they are on a wait list for funding. Any wait listed applicant that is eventually awarded funds will be offered the opportunity to register at the early registration rate, therefore, please do not register for the conference if your attendance is fully dependent on being awarded a travel fellowship as any cancellation of an applicant's registration will be subject to the full regular registration cancellation policy.
Funded applicants will be required to present evidence of their eligibility status (such as student identification card) when signing in daily to record their attendance. In all cases, funds will be mailed to funded applicants after the conference per the details noted in Eligibility Requirements above.
- top -
Contacts
Questions regarding travel fellowships should be addressed to: This email address is being protected from spambots. You need JavaScript enabled to view it.
The information on this page is subject to change without notice, and all changed information will be considered final for the purposes of awarding and funding GLBIO 2023 Travel Fellowships.
 Oana Carja
Oana Carja
USA
Tuesday, May 14, 2024 09:00
Dr. Carja works to quantitatively understand the evolutionary architecture of intelligent, collective systems, using the tools of dynamical systems, network theory, population genetics, machine learning and statistical inference, and widely available, yet underused, datasets.
Topological puzzles in biology: how shape and structure control a system's evolution
Understanding how the spatial arrangement of a population shapes its evolutionary dynamics has been of long-standing interest in evolutionary biology. However, most previous work assumes very regular symmetric structures, such as a small number of demes connected by migration corridors. This encoded regularity makes it hard to link these models to the influx of spatially resolved biological datasets that are revolutionizing molecular biology. In this talk I will explore how to think about and understand the role of complex spatial topologies in shaping a system's evolutionary dynamics and discuss the challenge of finding the right spatial geometric representations that can both capture the complexity of spatial structure in emerging datasets, but also allow for mathematical modeling tractability and interpretability.
- top -
 Mingfu Shao
Mingfu Shao
USA
Tuesday, May 14, 2024 13:30
Mingfu Shao is an assistant professor in the Department of Computer Science and Engineering, The Pennsylvania State University. He earned his PhD in Computer Science from École Polytechnique Fédérale de Lausanne (EPFL), Switzerland, in 2015. After that he pursued postdoctoral training as a Lane Fellow at Carnegie Mellon University prior to joining Penn State in 2018. Mingfu and his research group work on a variety of topics in algorithmic sequence analysis. They have been developing methods for assembling full-length transcripts from various RNA-seq data, including Scallop (for Illumina short-reads RNA-seq data), Scallop2 (for multi-end single-cell RNA-seq data), TERRACE (for assembling circular RNAs), and Aletsch (for assembling multiple RNA-seq samples). Mingfu’s group also works on designing efficient hashing and seeding algorithms for aligning and comparing sequencing data with high-error rates (e.g., error-prone long reads). Mingfu is a recipient of Dimitris N. Chorafas Foundation Award (2015) and NSF CAREER award (2022).
Transcript assembly for modern RNA-seq data
RNA-seq has been instrumental in studying gene activities. RNA-seq technologies evolve rapidly, enabling the profiling of various types of expressed RNAs with longer reads at single-cell resolution. These advancements bolster the broad use of RNA-seq but also bring new computational challenges for its analysis. In this talk, I will introduce our recent work in reconstructing full-length linear and circular isoforms from diverse RNA-seq data, a problem known as transcript assembly. I will highlight our new algorithms designed to capture the critical signals in the data, including paired-end information, barcode-linked reads, and multiple samples, thereby enhancing assembly accuracy. I will also discuss major (human) transcriptome annotations and their impact on transcript assembly.
- top -
 Nathan Clark
Nathan Clark
USA
Wednesday, May 15, 2024 09:00
Dr. Nathan Clark is an Associate Professor in Biological Sciences at the University of Pittsburgh. He trained in Genome Sciences at the University of Washington under Dr. Willie Swanson and later at Cornell with Dr. Charles Aquadro. His lab, which opened in 2012, studies the genetics of adaptive evolution, during which species adopt novel traits to overcome challenges. With the goal of identifying specific genetic elements responsible for adaptive change, his lab has developed computational methods used around the world for comparative genomics, Evolutionary Rate Covariation and RERconverge. The lab applies these methods to biological systems including eyesight, deepwater diving, and lifespan, among others.
Dr. Clark has created and led local seminar groups focused on building community in evolutionary genetics, and through training and education he works to increase diversity in science. He serves on the editorial board of Evolution, and in 2019, he received the Senior Vice Chancellor's Distinguished Research Award.
History Repeats Itself: Genetic changes underlying convergent phenotypic traits are revealed through comparative genomics
A combination of comparative genomics and phylogenetics is increasingly being used to assign genotype to phenotype through an approach called PhyloG2P. PhyloG2P methods search for positive selection, shifts in evolutionary rate, or pseudogenization associated with the branches over which a specific convergent phenotype evolved, such as long lifespan or loss of eyesight. These approaches have identified new genes and regulatory regions controlling traits important to fitness and health.
- top -
 Sarah Bagby
Sarah Bagby
USA
Wednesday, May 15, 2024 13:30
Sarah Bagby earned bachelor's degrees in biochemistry, chemistry, and philosophy at the University of Chicago, then studied physiology at Oxford as a Marshall Scholar. She returned to the US to earn her PhD in biology at MIT, working both on RNA biochemistry at the origins of life, in Dave Bartel’s lab, and on cyanobacterial physiology, with Penny Chisholm. In her postdoctoral research in Dave Valentine's lab at UC Santa Barbara she studied microbial metabolism at marine hydrocarbon seeps and spills. She is now an assistant professor in the Department of Biology at Case Western Reserve University, where her lab focuses on understanding how soil microbes balance their nutrient and energetic needs and how microbes acclimate and adapt to change in their environments. She sits on the executive committee of the EMERGE Biology Integration Institute.
Stable States in an Unstable Landscape
Soil is abundant, complex, critical to our food supply, and a massive reservoir of biodiversity. It is also changing fast. In the rapidly-warming boreal latitudes, permafrost thaw completely remodels soil chemistry, driving ecological succession and liberating huge stores of soil carbon---shifts whose climate impacts (e.g., will permafrost peatlands be net sources or net sinks of methane and CO2?) are not yet well understood. A critical gap in our ability to build predictive models of these globally significant ecosystems' climate outputs is our lack of understanding of how, exactly, peatland soil microbiomes respond to the coupled changes that accompany warming. Long-term ecological research in a model permafrost peatland system, northern Sweden's Stordalen Mire, enables us to ask this important question. At the macroscopic level, we observe rapid habitat change across the site's thaw gradient, and previous work has established that each distinct habitat along the gradient has its own characteristic plant and microbial communities. We asked whether these communities exhibit gradual or concerted change in a seven-year in situ metagenomic and metatranscriptomic survey. Through the lenses of network analysis, ecological assembly modeling, and multi-omic genome-resolved metabolic inference of carbon (C), nitrogen (N), and sulfur (S) cycles, we sought to identify drivers of soil microbiome change in a period of rampant macroscopic change. Remarkably, we found virtually no evidence of within-habitat change over time, instead identifying candidate mechanisms that may buffer these systems' community structure, assembly processes, and metabolic processes against change---until the buffer capacity is exceeded. I will discuss our analyses and what they mean for the prospect of decadal-scale predictive modeling of the climate consequences of permafrost thaw in Stordalen and other boreal peatlands.
- top -
 Daniel Lobo
Daniel Lobo
USA
Thursday, May 16, 2024 09:00
Dr. Daniel Lobo is an Associate Professor in the Department of Biological Sciences at the University of Maryland, Baltimore County and Member of the Greenebaum Comprehensive Cancer Center and the Center for Stem Cell Biology & Regenerative Medicine at the University of Maryland, School of Medicine. Dr. Lobo received his Ph.D. from the University of Malaga in Spain before completing his postdoctoral training at Tufts University in Massachusetts. Dr. Lobo’s lab in Systems Biology aims to understand, control, and design the dynamic regulatory mechanisms governing biological growth and form. To this end, his group combines molecular methods with computational approaches towards the reverse-engineering of mechanistic models from biological data and the automatic design of regulatory networks for specific functions. They seek to discover the mechanisms of development and regeneration, find therapies for cancer and other diseases, and streamline the application of systems and synthetic biology. Dr. Lobo has received several awards for his research, including the NIH Outstanding Investigator Award, the PhRMA Foundation Research Starter Award, and the UMBC Early Career Faculty Excellence Award. His work on Systems Biology has attracted widespread media coverage including PBS, Wired, TechRadar, and Popular Mechanics.
Inference and design of gene regulatory mechanisms for temporal and spatial phenotypes
The mechanisms controlling the formation of temporal and spatial patterns involve complex feedback loops, non-linear interactions, and emergent cellular behaviors. They are thus a challenge to infer from dynamic phenotypes as well as to design for bioengineering applications. In this talk I will present our bioinformatics and systems approach to automatically infer and design dynamic regulatory mechanisms. We demonstrated this methodology by mechanistically understanding whole-body multicellular dynamics, synthetic spatial patterns, and cancer clinical data.
- top -
 Andreas Pfenning
Andreas Pfenning
USA
Thursday, May 16, 2024 16:00
Andreas Pfenning is an Associate Professor in the Computational Biology Department in the School of Computer Science with a courtesy appointment in the Department of Biological Sciences at Carnegie Mellon University and he is a member of the Neuroscience Institute. Previously, he completed a joint postdoctoral position with Prof. Manolis Kellis in Computer Science at MIT (CSAIL) and Prof. Jesse Gray in the Genetics Department at Harvard Medical School. Andreas has a PhD in Computational Biology and Bioinformatics from Duke University where he worked with Prof. Erich D. Jarvis in neurobiology and Prof. Alexander Hartemink in Computer Science. Before that, Andreas obtained his undergraduate degree in computer science at Carnegie Mellon University.
The Pfenning lab studies how genetic and epigenetic variation influences complex neural traits. Their research has been published in top venues, including a series of Science articles applying comparative genomics to study genetic variation across species and within the human population. Andreas has won several additional accolades including a Sloan Foundation research fellowship, an NSF Career Award, and an American Federation for Aging Research grant. He also received an Avenir award, a grant given by the National Institute of Drug Abuse to fund forward-thinking research by new investigators in substance use disorders.
Linking Genetics to Behavior in the Evolution of Vocal Learning
Vocal production learning is a convergently evolved trait in vertebrates. To identify brain genomic elements associated with mammalian vocal learning, we applied neural network approaches to integrate genomic, anatomical and neurophysiological data from the Egyptian fruit-bat with analyses of the genomes of 215 placental mammals. In parallel, we applied single single cell genomics to the zebra finch brain to dissect out differences in gene regulatory patterns associated with vocal learning brain regions. Our analysis across mammals and birds implicates long range projection neurons in the evolution of vocal learning behavior.
- top -
Links in this page: General Submission Information | Full Papers for Oral Presentation | Abstracts for Oral Presentation or Poster | Questions
General Submission Information
Submissions are invited for full papers, oral presentation abstracts, and posters at the 16th Great Lakes Bioinformatics Conference, an official conference of the International Society for Computational Biology. This is an invitation to scientists and professionals working in the fields of bioinformatics and computational biology to submit high quality original research papers for presentation at GLBIO 2024. Authors of papers and oral abstracts that are accepted to the conference will present their work at the conference (longer talks for full papers, shorter talks for the abstracts). Accepted full papers will be considered for inclusion in a special issue of Bioinformatics Advances.
A wide definition and inclusion of bioinformatics/computational biology will be considered, and topics of interest include the following:
- Algorithms & Machine Learning
- Bioinformatics Education
- Biostatistics
- Cheminformatics
- Clinical & Health Informatics
- Databases, Ontologies & Biocuration
- Disease Models & Molecular Medicine
- Epidemiology & Biodiversity
- Evolutionary, Comparative Genomics & Phylogenetics
- Gene Regulation & Transcriptomics
- Genome Informatics
- Image Analysis
- Macromolecular Structure & Function
- Metagenomics
- Microbiome Informatics
- Network Biology
- Proteomics & Metabolomics
- Sequence Analysis
- Synthetic Biology
- Systems Modeling
- Text Mining & Natural Language Processing
- Visualization
Key dates: Click here
Special Sessions: Authors can request consideration by special sessions in Easychair. Please carefully review the information below before submitting in Easychair.
Sessions that will consider full Papers and Abstracts
Sessions that will consider Abstracts only
Submit Paper or Abstract
- top -
Full Paper for Oral Presentation
For a variety of reasons, ISCB/GLBC strongly prefers that scientific research accepted for oral presentation be presented in-person at the conference venue. We understand that some presenters will have valid reasons to avoid in-person attendance. ISCB will grant remote presentation options for reasons associated with maternity/paternity leave, care for a family member, personal/medical disability, sickness, financial hardship, or potential visa problems. If your research is accepted for oral presentation and you are unable to present in person, ISCB/GLBC requires notification at the time of acceptance and no later than April 12, 2024. You will be asked during your confirmation of participation to confirm your in-person participation. If unable to participate you will need to request a waiver by writing This email address is being protected from spambots. You need JavaScript enabled to view it.
The Bioinformatics Advances Journal has committed to considering publication of a special issue for peer-reviewed articles from the conference.
In addition to the standard submission information, authors are expected to submit a PDF of the written article following the preparation guidelines as outlined by Bioinformatics Advances
Article submissions will close February 5, 2024, initial review will be done and authors will have an opportunity to update and prepare their final manuscripts. The final date for updates will be March 8, 2024. Bioinformatics Advances will have the opportunity to review the papers and provide comments. We will give authors one month to reply to comments in order to have the list of accepted articles at the end of March. Outstanding articles will be selected for featuring talks.
Authors are responsible for all publishing fees.
Submit Paper or Abstract
- top -
Abstracts for Oral Presentation or Poster
Abstracts for Oral Presentation submissions to GLBIO 2024 should be approximately 400 words and have no figures. Adding references to already published papers is recommended where appropriate. Submission deadline in Easychair is March 4, 2024. Late Breaking Poster submission deadline is April 14.
For a variety of reasons, ISCB/GLBC strongly prefers that scientific research accepted for oral presentation be presented in-person at the conference venue. We understand that some presenters will have valid reasons to avoid in-person attendance. ISCB will grant remote presentation options for reasons associated with maternity/paternity leave, care for a family member, personal/medical disability, sickness, financial hardship, or potential visa problems. If your research is accepted for oral presentation and you are unable to present in person, ISCB/GLBC requires notification at the time of acceptance and no later than April 12, 2024. You will be asked during your confirmation of participation to confirm your in-person participation. If unable to participate you will need to request a waiver by writing This email address is being protected from spambots. You need JavaScript enabled to view it.
Any abstracts submitted after the deadline will be included only at the discretion of the conference chairs, and will be eligible for poster presentations only. Please also note that we can only allow one abstract per presenting author.
In-Person Poster Hall: When preparing accepted posters please note that your printed poster should not exceed the following dimensions: 46 inches wide by 46 inches high. There will be 1or 2 poster per side on the each poster board. Additionally, we ask that you also follow the Virtual Poster Hall information below in addition to your in-person presentation. This ensures all posters are viewable to both in-person and virtual attendees thus providing more exposure to your work.
Virtual Poster Hall: When preparing accepted posters please note that your poster will need to be uploaded via the virtual platform portal in two formats - a PDF of the poster and a 5-7 minute flash talk video not to exceed 100MB. Poster portal information will be sent to all approved posters closer to the conference and after the notification deadline. All presentations MUST be uploaded April 24-May 6*. More specific dates will be given closer to the conference.
*Late Breaking posters will have a very short turn-around.
Submit Paper or Abstract
- top -
Questions
For any GLBIO questions, please contact: Bel Hanson, ISCB Director of Operations and Programs, at This email address is being protected from spambots. You need JavaScript enabled to view it.
- top -
Links within this page: Customized workflows for the interactive visualization of large-scale biological networks: A Python-based framework | Beyond normalization: Incorporating scale into high-throughput sequencing analysis | Student-Focused Session: Navigating In-Person Conferences | Applications of Generative AI, LLMs, and Traditional Machine Learning for Personalized Medicine and Drug Discovery
Customized workflows for the interactive visualization of large-scale biological networks: A Python-based framework
Monday May 13, 2024 in Room 328 of the Mellon Institute at CMU08:00 AM - 12:00 PM
Networks are extremely useful tools for analyzing biological systems, and their effective visualization is crucial to properly interpreting and communicating their message. In large-scale networks with thousands of nodes and tens of thousands of edges, hand-tailoring the visualization is often impractical, therefore, it is hard to avoid a common pitfall of network visualization – the so-called “hairball network.” Interactive visualizations in which users can zoom in/out, pan, highlight, annotate, search, and select portions of the network offer a feasible middle ground that can help alleviate this challenge. Current freely available software packages and tools each lend some of their functionality to addressing parts of this task; however, end-to-end visualization workflows that integrate these individual parts are lacking, and knowledge thereof is sparse and not centralized. Our aim with this tutorial is to create working knowledge of the building blocks of interactive network visualizations, and to enable users to bring them together to design, create and deploy their own interactive network visualizations. Our tutorial will integrate elements from a lean set of tools (Gephi, NetworkX, Pajek, Bokeh, and Streamlit) in a Python-based framework, and use Google Colab for the majority of coding tasks.
Capacity: 40
Schedule
8:00 am – 10:00 am
Part I: To begin is half the work: A Pythonic preamble to preparing your network for Gephi
Having the right input is crucial to maximally leveraging the capabilities of Gephi. This portion will include a hands-on tutorial in Google Colab on how to generate multi-feature node and edge files for highly customized visualizations.
Part II: Visualizing your networks in Gephi – using key common and advanced functionalities for interactive visualizations
This portion of the tutorial will include an in-depth walkthrough of the visualization process with a focus on using Data Laboratory features that will enable downstream interactive visualizations. It will also include some helpful tips on layout choices and basic network analyses.
10:00-10:15 am
Coffee break
10:15 am - 12:00 pm
Part III: Using the Gephi output for inter-operable interactive visualizations
This portion of the tutorial will focus on using Bokeh to create interactive versions of the Gephi-generated networks and adding additional features such as dropdown menus and interaction events (e.g., mouse click and hover events) via custom JavaScript callbacks.
Part IV: Using Streamlit to create and deploy interactive network visualization applications
Building an interactive application with your networks can boost the visibility of your research. This portion of the tutorial will focus on designing and deploying a Streamlit web app of your interactive network visualizations.
Organizers
Arda Halu, PhD, Instructor in Medicine and Associate Scientist, Channing Division of Network Medicine, Brigham and Women’s Hospital, Harvard Medical School
Prerequisites
- Working knowledge of Python
- Gephi (installed locally on the participant's laptop)
- Access to Google Colab (requires a Google account)
- or -
The following Python libraries installed locally:
- Pandas
- NetworkX
- Bokeh
- Streamlit
- top -
Beyond normalization: Incorporating scale into high-throughput sequencing analysis
Monday May 13, 2024 in Room 348 of the Mellon Institute at CMU08:00 AM - 12:00 PM
In this workshop, we discuss a novel modeling framework for high-throughput sequencing (HTS) data that moves away from normalization-based analyses and addresses some critical limitations with normalization-based approaches. Many tools for analyzing sequence count data use some form of statistical normalization in an attempt to remove the non-biological variation in sequencing depth. However, simply removing the non-biological variation is not sufficient to recover biological insights from these data because the unmeasured variation in scale (i.e., size) is often important. Recently, we have shown that many normalizations make some identifying assumption about the scale of system in order to perform many common modeling tasks for HTS data (Nixon et al., 2023). Moreover, we found that even slight error in these assumptions could lead to a troubling phenomena called unacknowledged bias: statistical estimates are biased but this bias is not considered in corresponding uncertainty estimates (e.g., confidence intervals or credible sets). In the context of HTS analyses, we found this unacknowledged bias could lead to false positive rates as high as 80% and false negative rates as high as 86%. To address these limitations, we developed Scale Simulation Random Variables (SSRVs) which use a generalization of normalizations which we call scale models (Nixon et al., 2023). In contrast to normalizations, scale models make explicit, transparent, and modifiable assumptions about the system scale.
In this workshop, we will discuss the pitfalls associated with normalization-based analyses and demonstrate how incorporating scale uncertainty can address some of these limitations. Going further, we will show how scale can be incorporated into many analyses such as differential abundance, introduce a variety of easy-to-use software tools, and highlight recent modifications to the ALDEx2 software suite, which makes this software the first and only tool capable of considering scale uncertainty in analyses (i.e., considering uncertainty in the chosen normalization). Participants will gain an understanding of how the ALDEx2 software was modified to incorporate scale, including a discussion of the new functionality we have incorporated into the package to facilitate scale-based analyses. Furthermore, through guided analyses, we will show how incorporating scale can improve the flexibility, robustness, and reproducibility of HTS analyses and demonstrate how these tools can reveal novel biological insights by allowing analysts to create models that more faithfully reflect known patterns of scale variation (e.g., decreased microbial load after antibiotic treatment). Furthermore, workshop participants will have dedicated time to apply these tools to their own data with support from the workshop organizers.
Capacity: 30
Schedule
- Review of HTS data (e.g., collection and processing), common normalizations, their assumptions, and the concept of scale with a guided discussion/analysis of a previously published microbiome study.
- Introduction and guided discussion/analysis of incorporating scale uncertainty in analyses in ALDEx2 with application to the data sets from part 1.
- An introduction to Robust Differential Set Analysis including scale sensitivity analyses and the Log Fold Change Sensitivity Hypothesis Test with a guided DSA analysis to identify up- and down-regulated pathways in previously-published gene expression study.
- Dedicated time to discuss and apply learned concepts and tools to attendees’ own data.
Organizers
- Michelle P. Nixon, Ph.D: This email address is being protected from spambots. You need JavaScript enabled to view it.. Corresponding organizer
- Gregory Gloor, Ph.D: This email address is being protected from spambots. You need JavaScript enabled to view it..
- Justin D. Silverman, Ph.D., MD: This email address is being protected from spambots. You need JavaScript enabled to view it.
Prerequisites
Workshop participants are expected to have a basic knowledge of R. Installing the ALDEx2 (version 1.35.0 or higher), DESeq2, and fgsea packages prior to the workshop is recommended
References
- Nixon, M. P., Letourneau, J., David, L. A., Lazar, N. A., Mukherjee, S., and Silverman, J. D. (2023). Scale reliant inference. arXiv preprint arXiv:2201.03616.
- top -
Student-Focused Session: Navigating In-Person Conferences
Monday May 13, 2024 in Room 328 of the Mellon Institute at CMU1:00 PM - 5:00 PM
In this student-focused interactive half-day session we will have several activities designed to introduce trainees who may not have attended an in-person conference to the academic meeting environment. While the discussions will be geared toward first-time attendees, other students will be encouraged to attend not only to participate in the networking component of the session but to help impart any experiences from previous meetings. The day's events will include not only time for participants to hear from student-experts but also interact with other attendees in order to form connections that will be useful during the entire GLBIO meeting, and hopefully beyond.
Capacity: 50
Website: https://attending-conferences.github.io/glbio24.html
Schedule
| 13:00-13:45 |
Scientific Speed Dating |
| 13:45-14:00 |
Welcome and Introduction to Session |
| 14:00-14:45 |
Student Tips and Tricks Discussions |
| 14:45-16:00 |
Project Concept Ideation (submit topics here) |
| 16:00-16:45 |
Interactive Session on Science Communication featuring members of BioZone |
| 16:45-17:00 |
Closing remarks |
Organizers
- Dan DeBlasio, Assistant Teaching Professor, Carnegie Mellon University
- Raehash Shah, Undergraduate Student/Computational Biology Society President, Carnegie Mellon University
- Caleb Ellington, PhD Student/Graduate Student Association President, Carnegie Mellon University/CPCB
- Monica Dayao, PhD Student, Carnegie Mellon University/CPCB
Prerequisites
This session is open to trainees at any level (undergraduates, graduates, postdocs, etc.) who may not have attended an in-person meeting before, or have some tips and pointers that may be useful to others they want to share.
- top -
Applications of Generative AI, LLMs, and Traditional Machine Learning for Personalized Medicine and Drug Discovery
Monday May 13, 2024 in Room 348 of the Mellon Institute at CMU1:00 PM - 5:00 PM
As the amount of data increases, there is heightened complexity, complicating data science. Thus, a need exists for sophisticated technology that can address the changing landscape of data, harness inherent information and translate it to a tangible outcome.
The workshop will focus on applying new tools in the field of multi-omics, data integration, and artificial intelligence. For example, state-of-the-art software packages for precision medicine and large language models (LLMs) including OpenAI’s GPT-4 as well as other LLMs in the field. With greater understanding of existing tools and their use comes opportunity.
Capacity: 150
Schedule
Topic #1: Generative AI and LLMs
- Introduction to Generative AI and Open-Source LLMs
- Overview of NLP
- Applications of Generative AI and LLMs in Healthcare
- Medical Chatbots
- Medical Annotation Generator
- Omics Chatbot
Topic #2: AI for Personalized Medicine and Drug Discovery
- Introduction to Drug Response Prediction
- Model architectures
- Applications for Bulk and Single-cell RNA-seq
Organizers
- Gu, Quincy, Ph.D. in Bioinformatics and Computational Biology, Research Collaborator, Department of Neurology, Mayo Clinic
- Stover, Danielle Maeser, Ph.D. in Bioinformatics and Computational Biology, Senior Data Scientist, Humana This email address is being protected from spambots. You need JavaScript enabled to view it.
- Griffin, Nicole Maeser, M.S. in Bioinformatics and Computational Biology, Senior Bioinformatician, Contractor through Sault Tribe Inc. for the Centers for Disease Control and Prevention, Immunology and Pathogenesis Branch This email address is being protected from spambots. You need JavaScript enabled to view it.
- Patel, Ankush, Consultant, Mayo Clinic Platform, Mayo Clinic Department of Laboratory Medicine and Pathology; Consultant, Oncodea
Prerequisites
The workshops are relevant to all participants with an interest in understanding and/or adopting new technology to support healthcare (including undergraduates, graduate students, postdoctoral fellows, experimental researchers, industry professionals, etc.).
- top -






























 Sarah Bagby
Sarah Bagby





