Award Winners - ISMB 2014
Ian Lawson Van Touch Memorial Award
Outstanding Poster Award
F1000 Poster Award
RCSB PDB Poster Prize
Art & Science Award
Orienteering Winners
Ian Lawson Van Toch Memorial Award for Outstanding Student Paper
PP07 (PT) ExSPAnder: a Universal Repeat Resolver for DNA Fragment Assembly
Presenting author: Andrey D. Prjibelski, St. Petersburg Academic University, Russia
Additional authors:
Irina Vasilinetc, St. Petersburg Academic University, Russia
Anton Bankevich, St. Petersburg Academic University, Russia
Alexey Gurevich, St. Petersburg Academic University, Russia
Tatiana Krivosheeva, St. Petersburg Academic University, Russia
Sergey Nurk, St. Petersburg Academic University, Russia
Son Pham, University of California, San Diego, United States
Anton Korobeynikov, St. Petersburg Academic University, Russia
Alla Lapidus, St. Petersburg Academic University, Russia
Pavel Pevzner, University of California, San Diego, United States
Overview:
Next-generation sequencing (NGS) technologies have raised a challenging de novo genome assembly problem that is further amplified in recently emerged single-cell sequencing projects. While various NGS assemblers can utilize information from several libraries of read-pairs, most of them were originally developed for a single library and do not fully benefit from multiple libraries. Moreover, most assemblers assume uniform read coverage, condition that does nothold for single-cell projects where utilization of read-pairs is even more challenging. We have developed an exSPAnder algorithm that accurately resolves repeats in the case of both single and multiple libraries of read-pairs in both standard and single-cell assembly projects.
Poster E14
Modeling complex patterns of differential DNA methylation that strongly associate with gene expression changes
Christopher Schlosberg, Washington University in St. Louis, United States Nathan VanderKraats, Washington University in St. Louis, United States
Jeffery Hiken, Washington University in St. Louis, United States
Kilian Weinberger, Washington University in St. Louis, United States
Tao Ju, Washington University in St. Louis, United States
John Edwards, Washington University in St. Louis, United States
Short Abstract:
Establishment of specific patterns of DNA methylation is necessary for normal development, and aberrant methylation is frequently observed in cancer. Hypermethylation of CpG islands overlapping the transcription start site (TSS) downregulates tumor suppressor genes, thus promoting tumorigenesis. However, recent genome-wide mapping of methylation indicates only modest correlation between differential gene expression (DGE) and methylation, casting doubt on the importance of methylation in regulating DGE. In addition, complex patterns, such as CpG island-shore methylation and long hypomethylated domains, also correlate with DGE. We hypothesize that unbiased computational tools will better model complex patterns of methylation and capture strong associations between DGE and methylation. By representing methylation as continuous curves centered on a gene’s TSS and performing unsupervised clustering using Dynamic Time Warping, we enumerate complex, differential methylation signatures that highly correlate with DGE. We next trained a nearest neighbor classifier on examples of these significantly correlated signatures to identify genes that display both differential methylation and expression. Using data from the Human Epigenome Atlas, ENCODE, and eight breast cancer cell lines, our classifier significantly outperforms state-of-the-art Differentially Methylated Region (DMR)- and Support Vector Machine-based methods at identifying associated genes. By further analyzing these associated genes, we find methylation’s silencing mechanism may be signature-dependent. In breast cancer cells, we observe that methylation at the TSS does not affect transcriptional initiation, however, methylation proximal to the TSS may inhibit transcriptional elongation. The discovery of these potentially functional methylation changes will facilitate the identification of patients who may benefit from clinically-approved demethylating therapeutics.
Poster B27
Discovering co-variation and co-exclusion patterns in compositional data from the human microbiome
Emma Schwager, Harvard School of Public Health, United States
Curtis Huttenhower Harvard School of Public Health, United States
Uri Weingart, Harvard School of Public Health, United States
Timothy Tickle, Harvard School of Public Health, United States
Xochitl Morgan, Harvard School of Public Health, United States
Curtis Huttenhower, Harvard School of Public Health, United States
Short Abstract: Background: Compositional data, or data constrained to sum to a constant total, occur in many scientific areas. The non-independence of such data causes spurious correlations when standard covariance measures are applied, regardless of the similarity measure used. This problem has not yet been addressed in a way that generalizes to different similarity measures, nor for the high-dimensional measurements typical of modern biological data, including data from microbial community studies.
Results: We developed an approach to provide appropriate p-values for varied similarity scores between compositional measurements, which we call Compositionality Corrected by PErmutation and REnormalization (CCREPE). We assessed the false positive rate of CCREPE using synthetic datasets modeling a variety of realistic community structures, as well as comparing its performance and behavior with existing methods. We observed that CCREPE performs better in communities with greater evenness than in more skewed communities. We further applied the CCREPE procedure using a novel ecologically-targeted similarity score (the N-dimensional Checkerboard score) to 682 metagenomes from the Human Microbiome Project to determine significant co-variation patterns while avoiding spurious correlation from compositionality. Overall, the resulting network recapitulated the basic characteristics of earlier 16S-based networks, including little (<15%) between-site interaction and few "hub" microbes (scale-freeness).
Conclusions: These new methods will allow the derivation of significant co-variation networks from high-dimensional compositional data, particularly the detection of species and, eventually, sub-species level ecological interactions within microbial communities.
Poster A42
The Cure: Making a game of gene selection for breast cancer survival prediction
Benjamin Good, The Scripps Research Institute, United States
Karthik Gangavarapu, The Scripps Research Institute, United States
Salvatore Loguercio, The Scripps Research Institute, United States
Obi L. Griffith, Washington University School of Medicine, United States
Max Nanis, The Scripps Research Institute, United States
Chunlei Wu, The Scripps Research Institute, United States
Andrew I. Su, The Scripps Research Institute, United States
Short Abstract: Molecular signatures for predicting breast cancer prognosis could greatly improve care through personalization of treatment. Computational analyses of genome-wide expression datasets have identified such signatures, but these signatures leave much to be desired in terms of accuracy, reproducibility and biological interpretability. Methods that take advantage of structured prior knowledge show promise in helping to define better signatures but most knowledge remains unstructured.
Crowdsourcing via scientific discovery games is an emerging methodology that has the potential to tap into human intelligence at scales and in modes previously unheard of. Here, we developed and evaluated a game called The Cure on the task of gene selection for breast cancer survival prediction. Our central hypothesis was that knowledge linking expression patterns of specific genes to breast cancer outcomes could be captured from game players. We envisioned capturing knowledge both from the players prior experience and from their ability to interpret text related to candidate genes presented to them in the context of the game.
Between its launch in Sept. 2012 and Sept. 2013, The Cure attracted more than 1,000 registered players who collectively played nearly 10,000 games. Gene sets assembled through aggregation of the collected data clearly demonstrated the accumulation of relevant expert knowledge. In terms of predictive accuracy, these gene sets provided comparable performance to gene sets generated using other methods including those used in commercial tests. The Cure is available at http://genegames.org/cure/
Poster H20
GenTrAn: a new tool for de-novo transposon structural variant detection from single-end deep-sequencing data
Reazur Rahman, Brandeis University, United States
Yuliya Sytnikova, Brandeis University, United States
Nelson Lau, Brandeis University, United States
Short Abstract: Transposons are major structural variants (SVs) in animal genomes. In cancer and human biology, there is a need to determine new transposon SVs beyond the tremendous load of existing transposons (>45% of the human genome). Most current efforts to discover transposon SVs rely on Paired-End (PE) reads from genome deep-sequencing, but the greater costs of PE reads compared to Single-End (SE) reads (the standard form of genome deep-sequencing) motivated us to develop a new bioinformatics tool called GenTrAn (Genome Transposon Analyzer). By scanning SE read libraries with a hybrid approach of broad-level split-read mapping and then filtering with various quality criteria, GenTrAn discovers de-novo transposon SVs with high sensitivity and specificity. Importantly, the transposon SV sites that GenTrAn identifies display target site duplications indicative of a recent transposition event, and point to precise genomic coordinates that enable discrimination of SVs that disrupt coding gene exons versus less-disruptive intronic insertions.
We demonstrate the efficacy of our tool by discovering the genome-wide distributions of transposon SVs in four different Drosophila melanogaster cell lines. GenTrAn showed that transposon SV landscapes can be surprisingly diverse even in a natural cell line, and these SVs tend to avoid coding exons, yet prefer to insert near genes in intergenic regions. In addition, GenTrAn can measure the allele ratio of transposon SVs and all predicted SVs were successfully validated by genomic PCR. GenTrAn’s precision in transposon SV detection and feasibility to mine the more economical SE read libraries make this an attractive tool for genome diagnostics.
Poster P03
Biomedical Natural Language Figure Processing Assisting High-Throughput Data Analysis
Hong Yu, UMass Medical School, United States
Short Abstract: An intelligent figure search engine will not only assist biocuration and allow individual biomedical researcher to access figures more efficiently from full-text biomedical articles, but also is an important step towards automatic validations of genome-wide high-throughput predictions. With more and more full-text biomedical articles becoming open access (the Berlin Declaration on Open Access to Knowledge in the Sciences and Humanities, the Bethesda Statement on Open Access Publishing, and PubMed Central), we are developing a figure search system (available at http://figuresearch.askhermes.org) that
integrates natural language processing, image processing, machine learning, and user interfacing “biomedical Natural Language figure Processing” approaches for intelligent biomedical figure search (iBioFigureSearch). Our iBioFigureSearch associates each figure with text that describes the content of the figure, summarizes the associated text, ranks figures by their importance, and integrates both image and text for improved information retrieval. We have evaluated iBioFigureSearch by both intrinsic and task-driven extrinsic evaluation and found that iBioFigureSearch improves user-centered information seeking.
Poster H33
VARANT: An Open Souce Variant Annotation Tool
Kunal Kundu, Tata Consultancy Services, United States
Steven Brenner University of California at Berkeley, United States
Rajgopal Srinivasan, Tata Consultancy Services Ltd, India
Uma Sunderam, Tata Consultancy Services Ltd, India
Sadhna Rana, Tata Consultancy Services Ltd, India
Ajithavalli Chellapan, Tata Consultancy Services Ltd, India
Short Abstract: We describe and present a comprehensive and easily extensible open source tool for Human Genome Annotation called VARANT, written in the Python programming language. While several tools for annotating variants are available, we believe that VARANT distinguishes itself by being fully open source, capable of using multiple processors/cores for speedy annotation and providing extensive annotation of UTR and non-coding regions in addition to the customary annotations of genes. An additional highlight of the tool is the ability to incorporate various inheritance models into the annotation process, which when coupled with phenotype information can be used to quickly generate a list of prioritized genes and variants. The tool has been successfully used to identify causal variants in rare immuno-disorders.
Poster L51
Structural Analysis and Remodeling of T-Cell Receptors
Thomas Hoffmann, Technische Universität München, Germany
Iris Antes, Technische Universität München, Germany
Short Abstract: T-cells play a major role in the adaptive immune response. T-cell receptor molecules (TCRs) distinguish between self-peptides and pathogenic peptides presented by major histocompatibility complex molecules (MHC) on cell surfaces and thus initiating an immune response. Structural prediction of TCR:peptide:MHC (TCR-p-MHC) complexes would allow a better understanding of this interaction and thus of the molecular basis of T-cell signaling, which is important for the development of immunotherapies and rational vaccine design. TCRs share a common structural scaffold, although the sequence variations in their variable domains can be high and the human TCR repertoire was estimated to be at least 10^8. Thus, due to genetic and structural diversities of the receptor, modeling of TCR structures and their TCR-p-MHC complexes is a challenging task. We investigated the structural characteristics of the TCR variable domains, consisting of two chains (alpha and beta). The analysis showed that the orientation of the TCR beta chain relative to the TCR alpha chain is dependent on the TCR type and that the differences can be described by a common center of rotation. Based on this analysis we developed a force field based prediction tool, which allows predicting the correct TCR geometry for at least 83% of the structures of our test set. In the presentation we will discuss the new methodology and its performance.
The Best Artwork Award - $200 Cash Prize
Hai Fang, University of Bristol, United Kingdom
Julian Gough, University of Bristol, United Kingdom
supraHex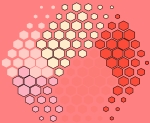 This artwork called ‘supraHex’ is inspired by the prevalence of natural objects such as a honeycomb or at Giant’s Causeway. supraHex has architectural design of a supra-hexagonal map: symmetric beauty around the center, from which smaller hexagons radiate circularly outwards. In addition to this architectural layout, supraHex also captures mechanistic nature of these objects: formation in a self-organising manner. For this, supraHex is able to self-organise the input data (eg transcriptome data). In doing so, genes with similar data patterns are clustered to the same or nearby nodes (hexagons). The map distance (the hexagon size) tells how far each node is away from its neighbors, thus characterising relationships between clustered genes. Based on this map distance, supraHex is also able to partition the map to obtain gene meta-clusters covering continuous regions, as colour-coded by the ‘potato-peach-tomato’ colormap. This artwork is generated by an open-source R/Bioconductor package ‘supraHex’ (http://supfam.org/supraHex).
This artwork called ‘supraHex’ is inspired by the prevalence of natural objects such as a honeycomb or at Giant’s Causeway. supraHex has architectural design of a supra-hexagonal map: symmetric beauty around the center, from which smaller hexagons radiate circularly outwards. In addition to this architectural layout, supraHex also captures mechanistic nature of these objects: formation in a self-organising manner. For this, supraHex is able to self-organise the input data (eg transcriptome data). In doing so, genes with similar data patterns are clustered to the same or nearby nodes (hexagons). The map distance (the hexagon size) tells how far each node is away from its neighbors, thus characterising relationships between clustered genes. Based on this map distance, supraHex is also able to partition the map to obtain gene meta-clusters covering continuous regions, as colour-coded by the ‘potato-peach-tomato’ colormap. This artwork is generated by an open-source R/Bioconductor package ‘supraHex’ (http://supfam.org/supraHex).
Bram Weytjens, Michael Crusoe, Sergio Pulido Tamayo, and Svetlana Vinogradova


