Award Winners
Ian Lawson Van Toch Memorial Award for Outstanding Student Paper | RCSB PDB Poster Prize | Bio-ontologies COSI | BioVis COSI | CompMS COSI | CAMDA Trophy Award for Best Presentation | EvolCompGen COSI | HiTSeQ Award | iRNA Award | MLCSB COSI | RegSys COSI | SysMod COSI | TransMed COSI
Award Winner:
Quentin Garrido, Université Gustave Eiffel, ESIEE Paris, LIGM, France
Visualizing hierarchies in scRNA-seq data using a density tree-biased autoencoder
Honorable Mentions:
Mohammadamin Edrisi, Rice University, United States
Phylovar: Towards scalable phylogeny-aware inference of single-nucleotide variations from single-cell DNA sequencing data
Bing-Xue Du, School of Life Sciences, Northwestern Polytechnical University, China
MLGL-MP: A Multi-Label Graph Learning Framework Enhanced by Pathway Interdependence for Metabolic Pathway Prediction
With Support From:


Nicola Bordin, University College London, London
AlphaFold2 reveals commonalities and novelties in protein structure space for 21 model organisms
COSI Awards
Bio-ontologies COSI
Best Poster
Michael Bradshaw, University of Colorado Boulder, United States
Identification of clusters containing future gene-to-phenotype relations across heterogeneous data sources
Best Talk
Frederic B. Bastian, University of Lausanne, SIB Swiss Institute of Bioinformatics, Switzerland
Creation and unification of development and life stage ontologies for animals
BioVis COSI
Best Long Abstract
Mark Keller, Harvard Medical School
Polyphony: an Interactive Transfer Learning Framework for Single-Cell Data Analysis
Best Poster
Camilo Valdes, Lawrence Livermore National Laboratory, United States
Microbiome Maps: Hilbert Curve Visualizations of Metagenomic Profiles
CompMS COSI
Best Talk
Daniela Klaproth-Andrade, Technical University of Munich, Germany
A genetic algorithm with deep learning-based guided mutations improves de novo peptide sequencing
Best Poster
Chathurani Ranathunge, Eastern Virginia Medical School, United States
promor: An Integrative Approach for Proteomics Data Analysis and Modeling
CAMDA Trophy Award for Best Presentation
Eunyoung Kim, Gwangju Institute of Science and Technology, South Korea
DeSIDE-DDI: Interpretable prediction of drug-drug interactions using drug-induced gene expressions
EvolCompGen Award
Talks
Winners
Taeyoung Kim, Giltae Song | Multi-modal Transformer based deep neural network for determining false positive structural variation calls
Joel Nitta, Eric Schuettpelz, Santiago Ramírez-Barahona, Wataru Iwasaki | An Open and Continuously Updated Fern Tree of Life (FTOL)
Honorable mentions
Mathieu Gascon, Yoann Anselmetti, Nadia El-Mabrouk | Non-binary Tree Reconciliation with Endosymbiotic Gene Transfer
Yuting Xiao, Maureen Stolzer, Dannie Durand | Domain Promiscuity Correlates with Rates of Domain Gain and Loss
Posters
Winner
Sungsik Kong, David Swofford, Laura Kubatko | Inference of phylogenetic networks from sequence data using composite likelihood
Honorable mentions
Ethan Wolfe, Joseph Burke, Stephanie Shames, Janani Ravi | Characterizing effector-metaeffector pairs in Legionella pneumophila
Chao Chun Liu, William Hsiao | Supervised machine learning reveals high efficacy of mobile elements to predict Salmonella outbreak linkage
Yiyan Yang, Xiaofang Jiang | Evolink: a Phylogenetic Approach for Rapid Identification of Phenotype-Genotype Associations in Large-scale Microbial Data
HiTSeQ Award
Best Talk
Rachael Aubin, University of Pennsylvania, United States
ConDecon: a clustering-independent method for estimating single-cell abundance in bulk tissues using reference single-cell RNA-seq data
iRNA Award
Best Posters
Christina Akirtava, Carnegie Mellon University, United States
Unraveling the roles of 5’ transcript leaders in gene regulation
Subhashis Natua, University of Illinois at Urbana-Champaign, United States
Post-transcriptional silencing of the nuclear poly(A) binding protein expression is essential for postnatal cardiac maturation and function
MLCSB COSI
Best Poster
Yusuf Roohani, Stanford University, United States
GEARS: Predicting transcriptional outcomes of novel multi-gene perturbations
NetBio COSI
Best Talk
Mikaela Koutrouli, Novo Nordisk Foundation Center of Protein Research, Denmark FAVA: High-quality functional association networks inferred from massive scRNA-seq and proteomics data
RegSys COSI
Best Talk
Jointly awarded to:
Pia Rautenstrauch, Max-Delbrück-Center for Molecular Medicine, Berlin, Germany “Liam tackles complex multimodal single-cell data integration challenges”
and
Anusri Pampari, Stanford University, United States “Base-resolution deep learning models of chromatin accessibility reveal combinatorial sequence motif syntax and regulatory variation”
SysMod COSI
1st Best Poster: Caroline I. Larkin, University of Pittsburgh, United States
A Mechanistic Model of Alphavirus Replication within a Mammalian Host Cell
2nd Best Poster: Kang Jin, Cincinnati Children's Hopsital Medical Center, United States
CellDrift: Inferring Perturbation Responses in Temporally-Sampled Single Cell Data
3rd Best Poster Award: Shaimaa Bakr, Stanford University, United States
Identifying key multifunctional components shared by critical cancer and normal liver pathways via sparseGMM
TransMed COSI
Best Talk (Best Oral Presentation)
Madison Darmofal, Memorial Sloan Kettering, Weill Cornell Graduate School, United States
Genome-Derived Diagnosis: Deep Learning Model for Tumor Type Prediction using MSK-IMPACT data
Yue Wang, University of Pittsburgh, United States
Perturbed Transcriptomic Analyses Identify Chemo-immunotherapy Synergisms to Shift Anti-PD1 Resistance in Cancer
Best Poster
Junjie Peng, University College London, United Kingdom
Patient-specific and disease-related determinants for cardiovascular disease (CVD) risk stratification in the APPLE (Atherosclerosis Prevention in Paediatric Lupus Erythematosus) clinical trial cohort
Jeppe Severens, Leiden University Medical Center, Netherlands
Joint genetics and transcriptomics based classification of acute myeloid leukaemia patients























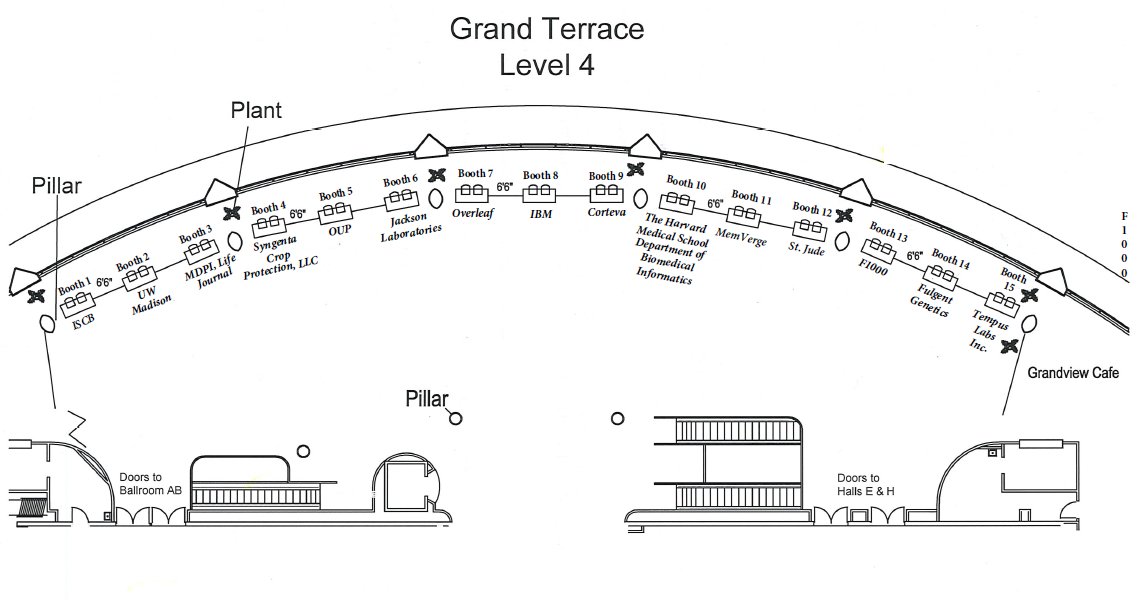

















 Please read the following information carefully and ensure that you have reviewed your Sponsor/Exhibitor Listing at
Please read the following information carefully and ensure that you have reviewed your Sponsor/Exhibitor Listing at 



