Art and Science Exhibition
ISMB 2014 brings together scientists from a wide range of disciplines, including biology, medicine, computer science, mathematics and statistics. In these fields people are constantly dealing with information in visual form: from microscope images and photographs of gels to scatter plots, network graphs and phylogenetic trees, structural formulae and protein models to flow diagrams; visual aids for problem-solving are omnipresent. Some of the works of the first such exhibition at the ISMB 2008 in Toronto combine outstanding beauty and aesthetics with deep insight that perfectly proves the validity of our approach or goes beyond the problem's solution. Others were surprising and inspiring through the transition from science to art, opening our eyes and minds to reflect on the work that we are undertaking.
The Art & Science Exhibition 2014 presents the artworks that have been generated as part of research projects. They are also soliciting images resulting from creative efforts that involve scientific concepts or employ scientific tools and methods.
The Art & Science Exhibition 2014 presents the artworks that have been generated as part of research projects. They are also soliciting images resulting from creative efforts that involve scientific concepts or employ scientific tools and methods.
Click image to download larger file
Art & Science Images | |
Forbes Burkowski University of Waterloo, Canada Molitor antifreeze H-bond “plates” are displayed here by generating a ruled surface to represent a quadrilateral formed by four protein backbone atoms surrounding a hydrogen bond. The plates provide a visual representation of the structural integrity of the protein’s interior. Coloration is related to the level of dehydron wrapping that protects the hydrogen bond (Fernández & Scott, Biophys. J., 2003). In this visualization, the architecture of the H-bond plates serves to describe a complex structural support system that is not readily apparent by observing the usual beta sheet representation. This is an antifreeze protein (PDB ID = 1EZG) from Tenebrio Molitor an insect that can survive on snow. Molitor is often referred to as a “snow flea” but is actually a spring-tailed insect that can move across damp snow by using the catapulting action of its tail. Since the insect is not warm-blooded, the antifreeze protein is necessary if it is to live in an icy environment. The display was generated by a Python script running within the UCSF Chimera environment. | 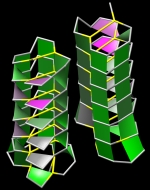 |
Forbes Burkowski University of Waterloo, Canada Twisted Ribonuclease H-bond “plates” are displayed here by generating a ruled surface to represent a quadrilateral formed by four protein backbone atoms surrounding a hydrogen bond. The plates provide a visual representation of the structural integrity of the protein’s interior. Coloration is related to the level of dehydron wrapping that protects the hydrogen bond (Fernández & Scott, Biophys. J., 2003). The protein is ribonuclease A (PDB ID = 1RHB) from Bos Taurus. The amphilicity of the alpha helix and the extreme twisting of the beta sheets provide a remarkable abstract sculptural design. The display was generated by a Python script running within the UCSF Chimera environment. | 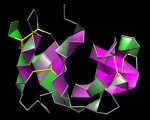 |
Forbes Burkowski University of Waterloo, Canada Malic enzyme H-bond “plates” are displayed here by generating a ruled surface to represent a quadrilateral formed by four protein backbone atoms surrounding a hydrogen bond. The plates provide a visual representation of the structural integrity of the protein’s interior. Coloration is related to the level of dehydron wrapping that protects the hydrogen bond (Fernández & Scott, Biophys. J., 2003). The protein is the human malic enzyme (PDB ID = 1PJ2). It is a tetrameric structure that has several helices and beta sheets. The H-bond plates serve to show the beta sheets as sheet-like structures and the overall symmetry of the structure adds to the visual appeal. The display was generated by a Python script running within the UCSF Chimera environment. |  |
Forbes Burkowski University of Waterloo, Canada Acostatin yin-yang H-bond “plates” are displayed here by generating a ruled surface to represent a quadrilateral formed by four protein backbone atoms surrounding a hydrogen bond. The plates provide a visual representation of the structural integrity of the protein’s interior. Coloration is related to the level of dehydron wrapping that protects the hydrogen bond (Fernández & Scott, Biophys. J., 2003). This protein is acostatin (PDB ID = 3C05) a dimeric disintegrin isolated from the Southern copperhead. The yin and yang configuration suggests the complementarity of contrary forces which in this case relates to the use of the venom as an anticancer agent. The display was generated by a Python script running within the UCSF Chimera environment. | 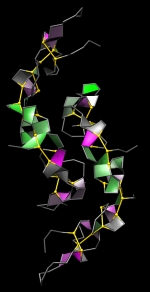 |
Frank Kramer University Medical Center Göttingen, Germany Michaela Bayerlova, University Medical Center Göttingen, Germany Three Interactomes This work illustrates a visual comparison of the Interactomes of three pathway databases (Reactome, PID, Pathway Commons), as compiled in my doctoral thesis. The Interactomes are the aggregation of all encoded regulatory interactions. The data has been downloaded from the database providers websites. It has been read into R via the rBiopaxParser, plotted using the Fruchterman-Reingold layout and beautified with gimp. |  |
Ludovic Antoine Alexandre University of Copenhagen, Denmark Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome A portrait representative of a Paleo-Eskimo individual of the Saqqaq culture from Greenland. Recent advances in ancient DNA research have allowed the sequencing of complete ancient genomes from short DNA molecules preserved in the fossil record for thousands, and sometimes even hundreds of thousands, of years. It was unknown, however, whether ancient epigenomes could be reconstructed. In this issue, the first ancient epigenome is reconstructed using DNA sequence information retrieved from the hairs of a 4000-yr-old Paleo-Eskimo individual. (Cover illustration by Nuka Konrad Gotfredsen, www.andala.dk. [For details, see Pedersen et al., pp. 454–466, Genome Research, 2014]) |  |
Roderic Guigo CRG, Spain Pinchas Akiva et al, Weizmann Institute of Science, Israel Rotem Sorek, Weizmann Institute of Science, Israel The phenomenon of Transcription-Induced Chimeras. Cover Image of Genome Research (January 2006): Transcription-Induced Chimeras (TICs) is a phenomenon that increases transcript and protein diversity in the human genome. Two consecutive genes are transcribed into a single pre-mRNA. Following splicing, the intergenic region is removed as an intron (upper right) and a chimeric mature mRNA is formed. This mRNA may be translated into a fused protein (lower left). The background figure is the mythological Chimera with a lion’s head, a goat’s body, and a serpent’s tail. (Cover art by Tali Cotter and Pinchas Akiva. [For details, see Akiva et al., pp. 30-36 and Parra et al., pp. 37-44, Genome Research, 2006]). |  |
Roderic Guigo CRG, Spain Hagen Tilgner, Stanford University, United States Chromatine is a key element in the genic regulation process The work that appears on the cover, with an illustration that could have been inspired by the Catalan artist Joan Miró, has been published in the journal Nature Structure & Molecular Biology. It is the result of a multidisciplinary collaboration between Bioinformatics, Genomics and Regulation of Gene Expression research groups at the Centre for Genomic Regulation ( CRG) affiliated to UPF. Roderic Guigó , one of the authors, said “it shows that ascertaining finding out the genetic code may not be enough for the difficulties we are finding in decoding the specific meaning of the genomic sequences, . Other factors, which until recently had been of little interest such as the architecture of DNA in chromatine, may play an important role in decoding its true biological meaning”. The published study describes how the structures responsible for packaging DNA in the nucleus also play an important role in interpreting the information contained in the genome. |  |
ALAA ABI HAIDAR University of Pierre and Marie Curie, France SMOKE The short clip and image series are not of smoke but of upside-down fluorescent paint dropped in fluorescent (quinine) tonic water under UV light. The diffusion of ink in water resembles smoke when flipped upside down. Both the clip and the images were shot with an SLR camera and a macro lens, however, this can be extended to UV light microscopy and telescopy. |  |
ALAA ABI HAIDAR University of Pierre and Marie Curie, France SMOKE The short clip and image series are not of smoke but of upside-down fluorescent paint dropped in fluorescent (quinine) tonic water under UV light. The diffusion of ink in water resembles smoke when flipped upside down. Both the clip and the images were shot with an SLR camera and a macro lens, however, this can be extended to UV light microscopy and telescopy. |  |
ALAA ABI HAIDAR University of Pierre and Marie Curie, France SMOKE The short clip and image series are not of smoke but of upside-down fluorescent paint dropped in fluorescent (quinine) tonic water under UV light. The diffusion of ink in water resembles smoke when flipped upside down. Both the clip and the images were shot with an SLR camera and a macro lens, however, this can be extended to UV light microscopy and telescopy. |  [Video] |
Georg Tremmel University of Tokyo, Japan Atsushi Niida, University of Tokyo, Japan Branching Evolutionary Process - Simulations (After several unsuccessful attempts of uploading a video, I'll try with only an image now. Please let me know, how I can transfer the video.) This video is an artistic interpretation of simulation data from our 'Agent-model based simulation of branching evolutionary process during tumorigenesis.' In these simulations, we aim to understand the mechanisms behind the emergence of intratumor heterogeneity. Cells are assumed to be agents, each fitted with an abstracted set of binary genes. The simulation is influence by various settings - growth rate, death rate, mutation rate, number of genes and driver genes. Our goal was to match the simulation data with the observed intratumor heterogeneity from patient samples. In this video the cells are cluster according to the similarity of their driver genes, each ray of the polar chart representing the average mutations within this cell cluster. Random values are used to define the initial and subsequent positions of the cells and their clusters. | 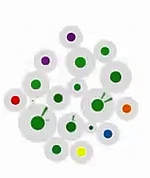 [Video] |
Ricardo Corral Corral Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, Mexico Fold Space This image shows an stylized embedding in two dimmensions of a 26-dimmensional protein fold space representation. A subset of 5,220 domains in CATH are represented as dots. Each dot is colored depending on one of the four main classes; mainly-alpha, mainly-beta, alfa-beta and few secondary structure. The position of each point depends only on structural features of the protein domain it represents. The structures are segregated in this space not only according to secondary structure content but also according to more sophisticated structural classifications. Regions in this space are expected to reveal functional similarities as well. For a recent discussion about the image of fold space as a framework to understand protein structure, funcion and evolution, refer to [1] [1] Rachel Kolodny, Leonid Pereyaslavets, Abraham O. Samson, and Michael Levitt, On the Universe of Protein Folds, Annu Rev. Biophys. 2013, 42:599-82 | 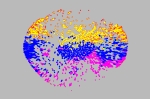 |
Hagen Tilgner Stanford University, United States Luisa Lente, Waverley St, Menlo Park, United States ENCODE project Cover of the "Genome Research", inspired by the work of Miró, where some of the findings of this study have been published. By Biocat A team of 442 scientists from around the world working with the ENCODE consortium has revealed that much of what had been called junk DNA in the human genome —which was believed to have no specific function— is actually a massive control panel with four million switches that regulate the activity of our genes. Without these switches, genes wouldn't work and mutations in these regions might lead to human disease. ENCODE is headed up by the US National Genome Research Institute (NHGRI) and the European Bioinformatics Institute (EMBL-EBI), with 32 other participating laboratories from the United Kingdom, the United States, Spain, Singapore, Japan and Switzerland. |  |
Karen Parker Moss Landing Marine Labs, United States Metabolite Transfer in a Marine Diatom This is a portrait of a one celled marine diatom. It depicts both passive and active transport of metabolites. On the left are the input metabolites and on the right are the output metabolites. In the center is the metabolic network. Small molecules like CO2 and O2 diffusive freely into and out of the diatom. Other molecules are transported by an active cell membrane transport structure. Diatoms produce about 20% of the oxygen in our atmosphere are about 50 microns in diameter. Diatoms have a beautiful and intricate silica outer covering called a frustule. Centric diatoms look like a pill box made out of rigid lace. In this painting the diatom is viewed from the side so it looks more rectangular than round. The design of this painting reflects the fact that the artist was a semiconductor engineer before she was a computational systems biologist. The diatom portrait almost looks like a semiconductor chip or a logic diagram with inputs and outputs. About the Artist Karen Parker is an oceanographer who built a metabolic network model of a marine diatom to investigate triacylglycerides. Her thesis won the 2014 Outstanding Thesis Award at San Jose State University. She has a strong interest in building metabolic models using Flux Balance Analysis (FBA) and kinetic modeling techniques. Prior to her career in computational systems biology, she worked as an engineer in the semiconductor business and received her undergraduate degree in Physics. | 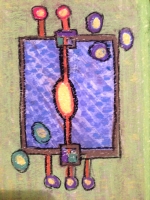 |
Karen Parker Moss Landing Marine Labs, United States Enzyme Activity This protein portrait depicts an ever moving, shape-changing protein immersed in a liquid cytosol landscape thick with molecules and activity. It is a snap shot of a landscape in motion. The small, fast metabolites are dwarfed by the large lumbering enzyme which has two small active reaction sites. In the background there are faint depictions of other proteins in action, suggesting the liquid flow of reactive interactions which make up a metabolic network. The watercolor pastel painting, which is part of a series, was copied onto note cards to share with friends and family who can relate more to art than science. The painting helped them relate to my work and to my passion for metabolic model building. About the Artist Karen Parker is a biological oceanographer who has built a metabolic network model of a marine diatom to investigate triacylglycerides. Triacylglycerides are important for both algal bio-fuel and omega-3 nutraceutical applications. Her thesis entitled the Metabolic Network Construction Based on the Genome of the Marine Diatom Thalassiosira Pseudonana and the Analysis of Genome-Wide Transcriptome Data to Investigate Triacylglyceride Accumulation, won the 2014 Outstanding Thesis Award at San Jose State University. She has a strong interest in building metabolic models with regulatory signaling elements using Flux Balance Analysis (FBA) and kinetic modeling techniques. Prior to her career in computational systems biology, she worked as an engineer in the semiconductor business and received her undergraduate degree in Physics. |  |
Max Nanis The Scripps Research Institute, United States Integrative Molecular Profile of Osteoarthritis Osteoarthritis (OA) is a prevalent joint disease that is characterized by the destruction of articular cartilage. We extracted total RNA, small RNAs and DNA from 8 normal and 10 OA donors. We then performed high-throughput transcriptome sequencing and analyzed DNA methylation. We identified 862 differentially expressed genes using the Tuxedo Pipeline. Using the Bioconductor package ChAMP to process HumanMethylation450 raw intensity data, we identified 2,833 differentially methylated sites, 170 of which were located in promoter regions. We discovered several upregulated miRs targeting aberrantly expressed genes in these pathways, such as miR-21, miR-10b, and miR-155. We also discovered several downregulated transcription factors, such as BHLHE40, FOSL2, and MAFF that are enriched for downregulated target mRNAs and are potential key regulators of this network. This molecular profile provides an integrated, systems-level understanding of the mechanisms of OA pathogenesis. The graphic shows the differentially expressed genes from our study separated by chromosome and directionality. They're horizontally positioned by each start position on their relative chromosome. The size of each symbol shows the log2FC value to indicate how differentially expressed each are while the color indicates up regulated (red) and down regulated (green). Symbol identifiers: genes are circles, CpG sites are squares and miRNA are triangles. | 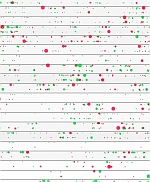 |
Max Nanis General Research Lab, United States BRCA2 The Sanger Series is a collection of drawings I create which honor the beauty of DNA and fluorescent emission from radioactive sequencing. A pioneer of DNA sequencing in the early 1970s, the work of Frederick Sanger has paved the way for modern high-throughput methods. The works are intended to convey a narrative of developmental passage. Mapping genes that are affected by non-fatal mutations or have great variance in expression levels often provide the viewer with a close relationship to their phenotypic expression. Using an algorithm I developed to model fluorescent radioactive emission output, the viewer is presented with a visually quantifiable drawing or surface. A, C, G, and T are identified by unique wave amplitudes to visually represent the human genetic code. The works transcend the viewer from an observer to a scientist. The patterns and forms of the drawings attempt to share the sequential information stored in the gene. Forcing viewers to question the purpose of the lines, they attempt to understand the importance of a single base. Through genomic patterns, the foreign language expresses itself in a manner that becomes legible to even an untrained eye. The viewer leaves with a clear visual understanding of the patterns which encode the assembly instructions for their own embodiment. The BRCA2 print was completed prior to the Myriad 2013 Supreme Court ruling that naturally isolated DNA is not patentable to be high-concept art to (potentially) mass distribute intellectual property. |  |
Guylaine Poisson University of Hawaii at Manoa, United States Céline Poisson, Université du Québec à Montréal, Canada Xavier Coulombe-Murray , Université du Québec à Montréal, Canada Anne Bergeron, Université du Québec à Montréal, Canada Mahdi Belcaid, University of Hawaii at Manoa, United States Aida Ouangraoua, Institut National de Recherche en Informatique et en Automatique, Lille, France Kyungim Baek, University of Hawaii at Manoa, United States Amalgamation of Information This figure addresses the challenge of representing structural information, self-alignments of tandem repeats, and conservation across species. Typographical superposition is used to merge three self-alignments from beta-propeller domains of Arabidopsis thaliana (ARA), Homo sapiens (HUM), and Neurospora crassa (NEU). Conservation is conveyed by recognizable letters, while elegant but cryptic symbols suggest diversity. The resulting alignment is broken into blocks, inverting some of them to mimic the structure of the WD40 motif. The superposition and inversion techniques are explained in the surrounding text by visual cues, and puzzles. | 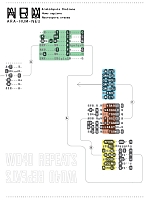 |
Chengsheng Zhu Rutgers University, United States Tom Delmont, Marine biological laboratory, United States Murat Eren, Marine biological laboratory, United States Function network of eight soil metagenomic samples We sequenced eight metagenomes of same soil samples under different treatments (control, ethanol, heavy metal and mercury; in duplicates). We annotated the functions represented in each metagenome sample directly from reads and visualized the overlaps. In this network figure, each metagenome sample node (Green: control; Yellow: ethanol; Blue: heavy metal; Red: mercury) connects to all its function nodes. Thus, common functions pull similar metagenome samples together while the unique functions separate them. As there are millions of functions, the visualization takes time, with more rare functions identified first, followed by more common, “network”, functions. As in life, however, only once all the functions are added in, do we see the real structure and relationships of our world. |  |
Lei Huang Houston Methodist Hospital Research Institute, United States biological networks This image contains two network figures of the accepted paper of ISMB 2014,'DrugComboRanker: Drug Combination Discovery Based on Target Network Analysis'. These two figures show the drug functional network with drug clusters and the disease dysfunctional signaling network, and different colors indicate different drug groups or gene clusters. In the latter figure, the node size indicates the degree of nodes. The drug functional network is actually a drug-drug network, and the disease dysfunctional signaling network is a protein-protein interaction network. |  |
Oleg Moskvin University of Wisconsin-Madison, United States Digital Life and Death You are swimming in the Ocean of Numbers. Those numbers represent hundreds of millions of genotype calls, millions of gene expression levels and ever-increasing number of clinical and other measurements taken from numerous cancer patients. You look at this Ocean and try to see, like in crystal ball, a reflection of future, a reflection of clinical outcome that is yet to come. And the crystal ball analogy fails right away because you don't see what you want to see at the surface of the Ocean... So you dive... And still don't see the answer... So you dive deeper, forgetting about the danger... This is Science. This is Art. This is Martial Art. You are fighting with Complexity. However, Her Highness Complexity is not willing to lose the game and easily reveal the internal structure hidden behind the apparent chaos of numbers. Your mind is hitting Complexity and being hit back over and over again. Every clash between the two gives a spark that usually lives a fraction of a second, and you still believe that one day, one of those sparks will condense into a small piece of stone in a huge brand new building of Personalized Medicine... |  |
Yuval Itan The Rockefeller University, United States A gene connectome A gene-specific connectome is the set of all biological distances and routes of all human genes from the specific gene of interest, identified by using a shortest distance algorithm such as in GPS navigation. Here we see the connectome of TLR3, a gene associated with herpes simplex encephalitis pathogenesis, depicted a medieval rose compass to reflect the navigation properties of both gene and traditional navigation. The image was generated with the help of Yael Pinchevsky Itan. |  |
Top


