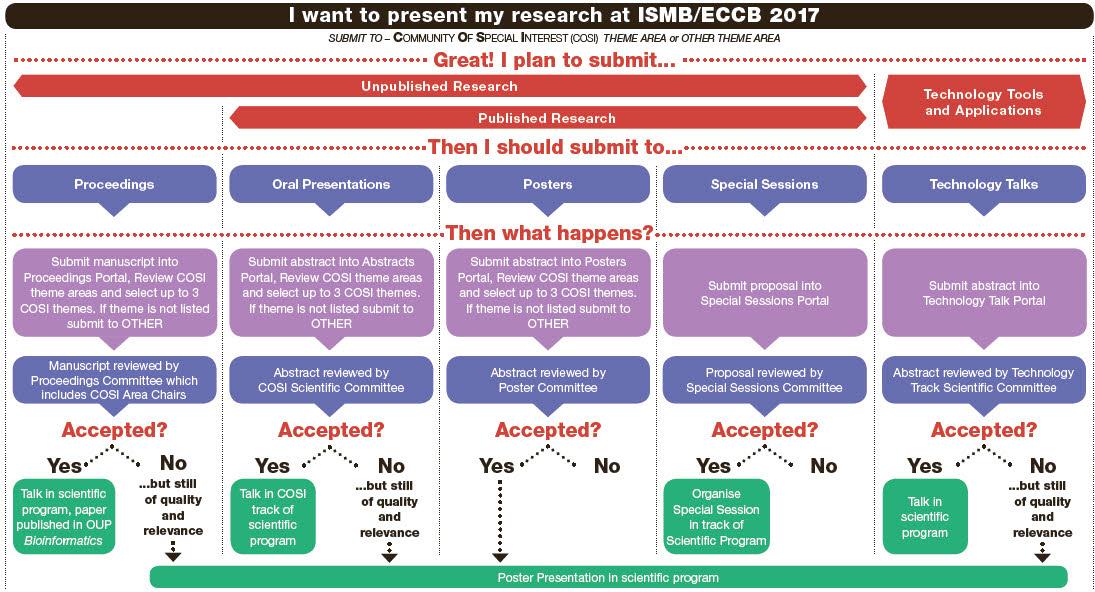
ISMB/ECCB 2017 Special Sessions
- SST01:Competency-based approaches to education and training in computational biology: bring your own educational challenge
- SST02:Critical Assessment of Metagenome Interpretation (CAMI)
- SST03:Computational Immune Oncology
- SST04:International Workshop on Machine Learning in Systems Biology (MLSB)
SST01: Competency-based approaches to education and training in computational biology: bring your own educational challenge
Saturday, July 22 2:00 pm – 6:00 pm Room: Meeting Hall V
Organizer(s):
Michelle Brazas, Ontario Institute for Cancer Research, Toronto, Canada
Catherine Brooksbank, EMBL-EBI, Cambridge, United Kingdom
Bruno Gaeta- University of New South Wales, Australia
Nicola Mulder- H3ABioNet, University of Cape Town, South Africa
Russell Schwartz- Carnegie Mellon University, United States
Lonnie Welch- Ohio University, United States
Presentation Overview:
Bioinformatics professionals work in a variety of settings, including core facilities, biological and medical research labs and software development organizations. The need for bioinformatics skills permeates academia, industry and the healthcare sector. The Curriculum Task Force (CTF) of ISCB’s Education Committee seeks to define curricular guidelines for those who educate or train bioinformatics professionals at all career stages. The task force has published a set of core competencies for bioinformatics professionals, which has been extensively revised in light of a thorough consultation process.
This highly practical and interactive workshop is aimed at education and training professionals in bioinformatics and other fields related to it. Participants will achieve the following outcomes:
- Gain insight into how competency-based approaches can be used to develop curricula, rapidly create cutting-edge training for established professionals, and empower their own team members to take ownership of their professional development
- Discover how this approach is being applied to bioinformatics and computational biology training programs in several different settings
- Create and/or refresh their own courses or curricula
- Provide input into the ISCB competency profile and contribute to its future development
This special session builds on a series of workshops run over the past two years in the US, Europe, Africa and Australia. We request that, should this application for a special session be accepted, that the timing is such that it does not overlap with any of the other education and training activities.
| 2:00 pm | Cath Brooksbank, EMBL-EBI and Lonnie Welch, Ohio University Status of the ISCB competency profile |
| 2:15 pm | Russell Schwartz, Carnegie Mellon University Keynote: Rethinking computational biology skills for molecular biology students |
| 2:30 pm | Bruno Gaeta, University of New South Wales Keynote: Developing the new generation of bioinformatics engineers |
| 2:45 pm | Nicola Mulder, H3ABioNet Applying competency-based approaches to a pan-African bioinformatics education initiative |
| 3:00 pm | Bruno Gaeta, UNSW Course and curriculum development breakout groups |
| 5:30 pm | Michelle Brazas, Ontario Institute for Cancer Research How to apply and improve curriculum and competency guidelines feedback |
Organizer(s):
Thomas Rattei, University of Vienna, Austria
Alexander Sczyrba, University of Bielefeld, CeBiTec, Germany
Alice McHardy, Helmholtz-Center Braunschweig, Germany
Presentation Overview:
The interpretation of metagenomes relies on sophisticated computational approaches such as short read assembly, binning and taxonomic classification. All subsequent analyses can only be as meaningful as the outcome of these initial data processing methods. Tremendous progress has been achieved during the last years. However, none of these approaches can completely recover the complex information encoded in metagenomes. Simplifying assumptions are needed and lead to strong limitations and potential inaccuracies in their practical use.
The accuracy of computational methods in metagenomics has so far been evaluated in publications presenting novel or improved methods. However, these snapshots are hardly comparable due to the lack of a general standard for the assessment of computational methods in metagenomics. Users are thus not well informed about general and specific limitations of computational methods. This may result in misinterpretations of computational predictions. Furthermore, method developers need to individually evaluate existing approaches in order to come up with ideas and concepts for improvements and new algorithms. This consumes substantial time and computational resources, and may introduce unintended biases.
We are in the process of tackling this problem with an initiative, aiming at the “Critical Assessment of Metagenome Interpretation” (CAMI). It evaluates methods in metagenomics comprehensively and objectively. The initiative supplies users with exhaustive quantitative data about the performance of methods in all relevant scenarios. It therefore guides users in the selection and application of methods and in their proper interpretation. Furthermore, CAMI provides valuable information to developers, allowing them to identify promising directions for their future work.
The session will focus on the CAMI initiative, which has successfully finished its first challenge. To continue and extend CAMI, we will bring together experienced developers of computational methods and researchers applying those in the major areas of metagenomics. During the session, we will present and discuss the results from the first CAMI challenge, and will determine the structure, targets and procedures of the next CAMI challenge, as well as the dissemination of its results. Microbiome research is currently one of the most dynamic fields in life sciences. It is of high relevance for (precision) medicine, global change research, microbial ecology, biotechnology and other areas.
| 10:00 am - 10:30 am | Shinichi Sunagawa, ETH Zurich; Keynote: "Metagenomics - from basics to applications in the human gut and ocean microbiome" |
| 10:30 am - 11:00 am | Alice McHardy, HZI Braunschweig: "Overview on CAMI, the initiative for Critical Assessment of Metagenome Interpretation" |
| coffee break | |
| 11:15 am - 11:40 am | Nils Willassen, UiT Tromso: "The ELIXIR Marine Metagenomics Use Case" |
| 11:40 am - 12:05 pm | Rob Finn, EBI Hinxton: "EBI metagenomics" |
| 12:05 pm - 12.30 pm | Christian Sieber, JGI: "Novel approaches for metagenomic assembly and binning" |
| lunch break | |
| 2:00pm - 3.00pm | Alex Sczyrba and colleagues: Practical demonstration of docker and bioboxes |
| 3:00 pm - 4:00 pm | Discussion on CAMI2 |
Organizer(s):
Jadwiga Bienkowska, Pfizer Oncology Research and Drug Development, San Diego, United States
Session Description:
In recent years cancer patients, who previously had very few treatment options and poor prognosis, have benefitted from new and lasting immuno-oncology therapies. These immuno-oncology treatments are now available to patients with advanced melanoma, and prostate and lung cancer, with other therapeutics and their combinations currently tested in extensive clinical trials. Particular advantage of these therapies is that they may work across a wide-range of cancer types. Therefore, Cancer Immuno Therapies are growing in importance to both healthcare providers and patients, and one that has already seen the success of large collaborative projects and new computational approaches benefiting drug discovery.
Session Description:
With the recent clinical success of checkpoint inhibitors (ipilimumab, pembrolizumab) generating durable responses in the clinic, immune modulation based therapy represents a promising modality for the treatment of cancer. The combination of immunotherapies with targeted therapies, including both small molecule and biotherapeutic, may drive this success further to those patients not responding to or relapsing with single agent immune therapies. Preclinical in vivo models for most immuno-oncology (IO) programs require the use of immunocompetent mice bearing syngeneic tumors. To facilitate model selection for use in preclinical efficacy studies, we characterized a panel of mouse tumor cell lines and syngeneic tumor tissues. Here we report the integrative analysis of gene expression and mutation landscape of these mouse tumor cells grown both in vitro and in vivo. Chromogenic immunohistochemistry (IHC) assays to identify and characterize infiltrating cell populations, including tumor infiltrating lymphocytes, myeloid cells as well as costimulatory and inhibitory markers, were developed and applied to these models.
Transcript expression data in general showed good agreement with the orthogonal IHC data for immune cell subsets. Known IO targets and immune cytolytic activity were up-regulated in the tumor compared to the matched in vitro cell pellet for solid tumors, indicating immune infiltration into the tumor. To further interrogate the immune subsets in the tumor microenvironment of syngeneic tumors, we developed a mouse immune cell-type specific gene signature and applied in silico deconvolution of the immune subsets using the recently published CIBERSORT method. In silico deconvolution and IHC analyses both revealed that these models display characteristics of low T-lymphocyte infiltration but relatively high myeloid cell infiltration. In addition to its high mutational load, the CT26 model also displayed the highest cytolytic activity among all models, in agreement with its known high immunogenicity. In vivo efficacy studies evaluating known IO therapy demonstrated that the in vivo response was correlated to genomic and IHC data. A detailed understanding of syngeneic models at both molecular and cellular level provides information for model selection, correlations with preclinical efficacy and will help translate preclinical findings to patient selection for clinical studies.
Session Description:
Epitopes that arise from a somatic mutation, so called neoepitopes, are now known to play a key role in cancer immunology and immunotherapy. Recent advances in highthroughput sequencing have made it possible to identify all mutations and thereby all potential neoepitope candidates in an individual cancer. It has however become evident that the vast majority of these neoepitope candidates do not induce a T cell response when tested in vivo or in vitro, i.e. they are not immunogenic. Especially in patients with a high mutational load, usually hundreds of potential neoepitopes are detected, highlighting the need to further narrow down this candidate list. Several studies have used different combinations of immunoinformatic tools such as MHC binding predictions to prioritize the initial set of neoepitopes candidates. The tools to use and thresholds to apply for this prioritization has so far been largely based on experience with epitope identification in other settings such as infectious disease and allergy. To establish the appropriate tools and thresholds in the cancer setting, we here curated a set of immunogenic neoepitopes from the published literature and performed detailed analyses to detect what features discriminate immunogenic neoepitopes from a background set of mutated peptides. We experimentally measured the HLA binding affinity of all curated immunogenic neoepitopes. In doing so, we aimed to identify the optimal affinity threshold to effectively identify immunogenic neoepitopes. As a next step, we sought to assess the added value of different immunoinformatics tools, including various HLA binding prediction algorithms, processing prediction, stability prediction, andimmunogenicity prediction, to most effectively detect immunogenic neoepitopes. The obtained results are now going to be used to facilitate the development of more accurate prediction algorithms.
Session Description:
Neoplastic cells reside within a complex tumor microenvironment consisting of tumorinfiltrating leukocytes (TILs) and non-hematopoietic stromal subsets that are necessary for tumor growth and survival. While flow cytometry and immunohistochemistry are commonly used to characterize tissue heterogeneity, the former requires cell dissociation, which can alter representation, while the latter is generally limited to one marker per section. Although single cell RNA sequencing has recently emerged as a powerful technology for defining novel cell subsets, it is currently impractical for large-scale analyses and cannot be applied to formalin fixed specimens. To complement these methods and to facilitate profiling of cellular heterogeneity in fresh, frozen, and fixed tissues, we introduced CIBERSORT, an “in silico flow cytometry” method for enumerating cell subsets of interest from gene expression profiles of intact bulk tumors.
In a pan-cancer analysis of nearly 6,000 human tumor samples, CIBERSORT revealed important new associations between TILs and clinical outcomes. In more recent work, we have extended this technique to accurately estimate cell type-specific gene expression profiles from tumor samples without the need for fluorescence-activated cell sorting.
Together, these methods comprise a versatile framework for digital cytometry with diverse applications in immuno-oncology, including identifying predictive and prognostic cellular biomarkers, and novel therapeutic targets.
Organizer(s):
Chloe-Agathe Azencott, MINES ParisTech, France
Magnus Rattray, University of Manchester, United Kingdom
Presentation Overview: (additional details available at: www.mlsb.cc)
Biology is rapidly turning into an information science, thanks to enormous advances in the ability to observe the molecular properties of cells, organs and individuals. This wealth of data allows us to model molecular systems at an unprecedented level of detail and to start to understand the underlying biological mechanisms. This field of systems biology creates a huge need for methods from machine learning, which find statistical dependencies and patterns in these large-scale datasets and that use them to establish models of complex molecular systems. MLSB is a scientific forum for the exchange between researchers from Systems Biology and Machine Learning, to promote the exchange of ideas, interactions and collaborations between these communities.
Motivation: Molecular biology and all the biomedical sciences are undergoing a true revolution as a result of the emergence and growing impact of a series of new disciplines and tools sharing the "-omics" suffix in their name. These include in particular genomics, transcriptomics, proteomics and metabolomics, devoted respectively to the examination of the entire systems of genes, transcripts, proteins and metabolites present in a given cell or tissue type.
The availability of these new, highly effective tools for biological exploration is dramatically changing the way one performs research in at least two respects. First, the amount of available experimental data is not a limiting factor anymore; on the contrary, there is a plethora of it. Given the research question, the challenge has shifted towards identifying the relevant pieces of information and making sense out of it (a "data mining" issue). Second, rather than focusing on components in isolation, we can now try to understand how biological systems behave as a result of the integration and interaction between the individual components that one can now monitor simultaneously (so called "systems biology").
Taking advantage of this wealth of "genomic" information has become a conditio sine qua non for whoever has the ambition to remain competitive in molecular biology and in the biomedical sciences in general. Machine learning naturally appears as one of the main drivers of progress in this context, where most of the targets of interest deal with complex structured objects: sequences, 2D and 3D structures or interaction networks. At the same time, bioinformatics and systems biology have already induced significant new developments of general interest in machine learning, for example in the context of learning with structured data, graph inference, semi-supervised learning, system identification, and novel combinations of optimization and learning algorithms.
| 8:40 AM-9:30 AM | Lineage estimation from single-cell RNAseq time-series |
| 10:00 AM-10:25 AM | Transcriptome-wide splicing quantification in single cells |
| 10:25 AM-10:50 AM | Gaussian processes for identifying branching dynamics in single cell data |
| 10:50 AM-11:40 AM | Data Integration in Computational Biology and Medicine: Current Progress and Future Directions |
| 11:40 AM-12:05 PM | Modeling Post-treatment Gene Expression Change with a Deep Generative Model |
| 2:15 PM-2:40 PM | Generative Learning of Dynamic Structures using Spanning Arborescence Sets |
| 2:40 PM-3:30 PM | Understanding and predicting drug efficacy in cancer: from machine learning to biochemical models |
| 3:30 PM-3:55 PM | Kernelized Rank Learning for Personalized Drug Recommendation |
| 3:55 PM-4:20 PM | Ask the doctor - Improving drug sensitivity predictions through active expert knowledge elicitation |